当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Diffusion-Based Generative Network for de Novo Synthetic Promoter Design
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2024-04-13 , DOI: 10.1021/acssynbio.4c00041 Jianfeng Lin 1 , Xin Wang 1 , Tuoyu Liu 2 , Yue Teng 2 , Wei Cui 1
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2024-04-13 , DOI: 10.1021/acssynbio.4c00041 Jianfeng Lin 1 , Xin Wang 1 , Tuoyu Liu 2 , Yue Teng 2 , Wei Cui 1
Affiliation
|
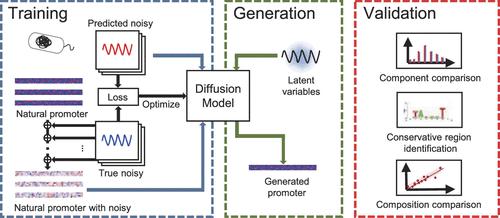
Computer-aided promoter design is a major development trend in synthetic promoter engineering. Various deep learning models have been used to evaluate or screen synthetic promoters, but there have been few works on de novo promoter design. To explore the potential ability of generative models in promoter design, we established a diffusion-based generative model for promoter design in Escherichia coli. The model was completely driven by sequence data and could study the essential characteristics of natural promoters, thus generating synthetic promoters similar to natural promoters in structure and component. We also improved the calculation method of FID indicator, using a convolution layer to extract the feature matrix of the promoter sequence instead. As a result, we got an FID equal to 1.37, which meant synthetic promoters have a distribution similar to that of natural ones. Our work provides a fresh approach to de novo promoter design, indicating that a completely data-driven generative model is feasible for promoter design.
中文翻译:

用于从头合成启动子设计的基于扩散的生成网络
计算机辅助启动子设计是合成启动子工程的主要发展趋势。各种深度学习模型已被用于评估或筛选合成启动子,但关于从头设计启动子的工作很少。为了探索生成模型在启动子设计中的潜在能力,我们在大肠杆菌中建立了基于扩散的启动子设计生成模型。该模型完全由序列数据驱动,可以研究天然启动子的本质特征,从而产生与天然启动子在结构和组成上相似的合成启动子。我们还改进了FID指标的计算方法,使用卷积层来提取启动子序列的特征矩阵。结果,我们得到的 FID 等于 1.37,这意味着合成启动子的分布与天然启动子相似。我们的工作为从头设计启动子提供了一种新的方法,表明完全数据驱动的生成模型对于启动子设计是可行的。
更新日期:2024-04-13
中文翻译:

用于从头合成启动子设计的基于扩散的生成网络
计算机辅助启动子设计是合成启动子工程的主要发展趋势。各种深度学习模型已被用于评估或筛选合成启动子,但关于从头设计启动子的工作很少。为了探索生成模型在启动子设计中的潜在能力,我们在大肠杆菌中建立了基于扩散的启动子设计生成模型。该模型完全由序列数据驱动,可以研究天然启动子的本质特征,从而产生与天然启动子在结构和组成上相似的合成启动子。我们还改进了FID指标的计算方法,使用卷积层来提取启动子序列的特征矩阵。结果,我们得到的 FID 等于 1.37,这意味着合成启动子的分布与天然启动子相似。我们的工作为从头设计启动子提供了一种新的方法,表明完全数据驱动的生成模型对于启动子设计是可行的。



























 京公网安备 11010802027423号
京公网安备 11010802027423号