当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Structure-Based Protein Assembly Simulations Including Various Binding Sites and Conformations
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-04-11 , DOI: 10.1021/acs.jcim.4c00212 Luis J. Walter 1 , Patrick K. Quoika 1 , Martin Zacharias 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-04-11 , DOI: 10.1021/acs.jcim.4c00212 Luis J. Walter 1 , Patrick K. Quoika 1 , Martin Zacharias 1
Affiliation
|
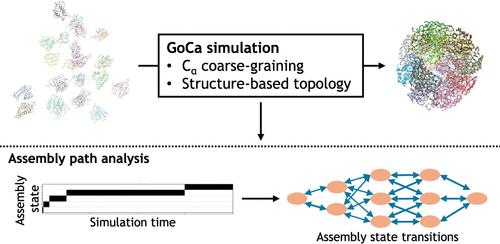
Many biological functions are mediated by large complexes formed by multiple proteins and other cellular macromolecules. Recent progress in experimental structure determination, as well as in integrative modeling and protein structure prediction using deep learning approaches, has resulted in a rapid increase in the number of solved multiprotein assemblies. However, the assembly process of large complexes from their components is much less well-studied. We introduce a rapid computational structure-based (SB) model, GoCa, that allows to follow the assembly process of large multiprotein complexes based on a known native structure. Beyond existing SB Go̅-type models, it distinguishes between intra- and intersubunit interactions, allowing us to include coupled folding and binding. It accounts automatically for the permutation of identical subunits in a complex and allows the definition of multiple minima (native) structures in the case of proteins that undergo global transitions during assembly. The model is successfully tested on several multiprotein complexes. The source code of the GoCa program including a tutorial is publicly available on Github: https://github.com/ZachariasLab/GoCa. We also provide a web source that allows users to quickly generate the necessary input files for a GoCa simulation: https://goca.t38webservices.nat.tum.de.
中文翻译:

基于结构的蛋白质组装模拟,包括各种结合位点和构象
许多生物功能是由多种蛋白质和其他细胞大分子形成的大型复合物介导的。实验结构测定以及使用深度学习方法的集成建模和蛋白质结构预测方面的最新进展导致已解决的多蛋白质组装体数量迅速增加。然而,大型复合体的组件组装过程却很少得到充分研究。我们引入了一种基于快速计算结构 (SB) 的模型 GoCa,它允许跟踪基于已知天然结构的大型多蛋白复合物的组装过程。除了现有的 SB Go̅ 型模型之外,它还区分了亚基内和亚基间的相互作用,使我们能够包括耦合折叠和结合。它自动解释复合物中相同亚基的排列,并允许在组装过程中经历全局转变的蛋白质的情况下定义多个最小(天然)结构。该模型已在多种多蛋白复合物上成功进行了测试。 GoCa 程序的源代码(包括教程)可在 Github 上公开获取:https://github.com/ZachariasLab/GoCa。我们还提供了一个网络源,允许用户快速生成 GoCa 模拟所需的输入文件:https://goca.t38webservices.nat.tum.de。
更新日期:2024-04-11
中文翻译:

基于结构的蛋白质组装模拟,包括各种结合位点和构象
许多生物功能是由多种蛋白质和其他细胞大分子形成的大型复合物介导的。实验结构测定以及使用深度学习方法的集成建模和蛋白质结构预测方面的最新进展导致已解决的多蛋白质组装体数量迅速增加。然而,大型复合体的组件组装过程却很少得到充分研究。我们引入了一种基于快速计算结构 (SB) 的模型 GoCa,它允许跟踪基于已知天然结构的大型多蛋白复合物的组装过程。除了现有的 SB Go̅ 型模型之外,它还区分了亚基内和亚基间的相互作用,使我们能够包括耦合折叠和结合。它自动解释复合物中相同亚基的排列,并允许在组装过程中经历全局转变的蛋白质的情况下定义多个最小(天然)结构。该模型已在多种多蛋白复合物上成功进行了测试。 GoCa 程序的源代码(包括教程)可在 Github 上公开获取:https://github.com/ZachariasLab/GoCa。我们还提供了一个网络源,允许用户快速生成 GoCa 模拟所需的输入文件:https://goca.t38webservices.nat.tum.de。



























 京公网安备 11010802027423号
京公网安备 11010802027423号