当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Discovery of Covalent Lead Compounds Targeting 3CL Protease with a Lateral Interactions Spiking Neural Network
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-03-23 , DOI: 10.1021/acs.jcim.3c01900 Zhihao Gu 1, 2 , Yong Yan 1 , Hanwen Liu 1 , Di Wu 1 , Hequan Yao 1 , Kejiang Lin 1 , Xuanyi Li 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2024-03-23 , DOI: 10.1021/acs.jcim.3c01900 Zhihao Gu 1, 2 , Yong Yan 1 , Hanwen Liu 1 , Di Wu 1 , Hequan Yao 1 , Kejiang Lin 1 , Xuanyi Li 1
Affiliation
|
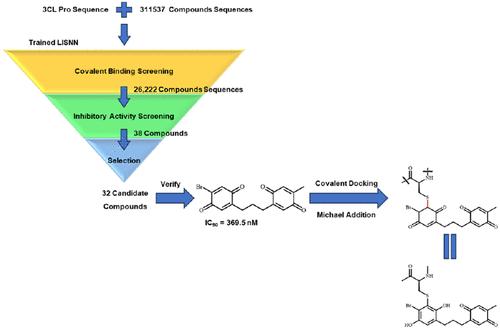
Covalent drugs exhibit advantages in that noncovalent drugs cannot match, and covalent docking is an important method for screening covalent lead compounds. However, it is difficult for covalent docking to screen covalent compounds on a large scale because covalent docking requires determination of the covalent reaction type of the compound. Here, we propose to use deep learning of a lateral interactions spiking neural network to construct a covalent lead compound screening model to quickly screen covalent lead compounds. We used the 3CL protease (3CL Pro) of SARS-CoV-2 as the screen target and constructed two classification models based on LISNN to predict the covalent binding and inhibitory activity of compounds. The two classification models were trained on the covalent complex data set targeting cysteine (Cys) and the compound inhibitory activity data set targeting 3CL Pro, respected, with good prediction accuracy (ACC > 0.9). We then screened the screening compound library with 6 covalent binding screening models and 12 inhibitory activity screening models. We tested the inhibitory activity of the 32 compounds, and the best compound inhibited SARS-CoV-2 3CL Pro with an IC50 value of 369.5 nM. Further assay implied that dithiothreitol can affect the inhibitory activity of the compound to 3CL Pro, indicating that the compound may covalently bind 3CL Pro. The selectivity test showed that the compound had good target selectivity to 3CL Pro over cathepsin L. These correlation assays can prove the rationality of the covalent lead compound screening model. Finally, covalent docking was performed to demonstrate the binding conformation of the compound with 3CL Pro. The source code can be obtained from the GitHub repository (https://github.com/guzh970630/Screen_Covalent_Compound_by_LISNN).
中文翻译:

发现具有横向相互作用尖峰神经网络的靶向 3CL 蛋白酶的共价先导化合物
共价药物表现出非共价药物无法匹配的优势,共价对接是筛选共价先导化合物的重要方法。然而,共价对接很难大规模筛选共价化合物,因为共价对接需要确定化合物的共价反应类型。在这里,我们建议利用横向相互作用尖峰神经网络的深度学习构建共价先导化合物筛选模型,以快速筛选共价先导化合物。我们以SARS-CoV-2的3CL蛋白酶(3CL Pro)为筛选目标,构建了两个基于LISNN的分类模型来预测化合物的共价结合和抑制活性。这两个分类模型在针对半胱氨酸(Cys)的共价复合物数据集和针对3CL Pro的化合物抑制活性数据集上进行训练,受到尊重,具有良好的预测精度(ACC > 0.9)。然后我们用 6 个共价结合筛选模型和 12 个抑制活性筛选模型筛选了筛选化合物库。我们测试了32种化合物的抑制活性,最好的化合物抑制SARS-CoV-2 3CL Pro,IC 50值为369.5 nM。进一步分析表明,二硫苏糖醇可以影响该化合物对3CL Pro的抑制活性,表明该化合物可能与3CL Pro共价结合。选择性测试表明,该化合物对3CL Pro具有优于组织蛋白酶L的良好靶点选择性。这些相关性分析可以证明共价先导化合物筛选模型的合理性。最后,进行共价对接以证明该化合物与3CL Pro的结合构象。源代码可以从 GitHub 存储库 (https://github.com/guzh970630/Screen_Covalent_Compound_by_LISNN) 获取。
更新日期:2024-03-23
中文翻译:

发现具有横向相互作用尖峰神经网络的靶向 3CL 蛋白酶的共价先导化合物
共价药物表现出非共价药物无法匹配的优势,共价对接是筛选共价先导化合物的重要方法。然而,共价对接很难大规模筛选共价化合物,因为共价对接需要确定化合物的共价反应类型。在这里,我们建议利用横向相互作用尖峰神经网络的深度学习构建共价先导化合物筛选模型,以快速筛选共价先导化合物。我们以SARS-CoV-2的3CL蛋白酶(3CL Pro)为筛选目标,构建了两个基于LISNN的分类模型来预测化合物的共价结合和抑制活性。这两个分类模型在针对半胱氨酸(Cys)的共价复合物数据集和针对3CL Pro的化合物抑制活性数据集上进行训练,受到尊重,具有良好的预测精度(ACC > 0.9)。然后我们用 6 个共价结合筛选模型和 12 个抑制活性筛选模型筛选了筛选化合物库。我们测试了32种化合物的抑制活性,最好的化合物抑制SARS-CoV-2 3CL Pro,IC 50值为369.5 nM。进一步分析表明,二硫苏糖醇可以影响该化合物对3CL Pro的抑制活性,表明该化合物可能与3CL Pro共价结合。选择性测试表明,该化合物对3CL Pro具有优于组织蛋白酶L的良好靶点选择性。这些相关性分析可以证明共价先导化合物筛选模型的合理性。最后,进行共价对接以证明该化合物与3CL Pro的结合构象。源代码可以从 GitHub 存储库 (https://github.com/guzh970630/Screen_Covalent_Compound_by_LISNN) 获取。



























 京公网安备 11010802027423号
京公网安备 11010802027423号