当前位置:
X-MOL 学术
›
Biotechnol. J.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Proteomic workflows for deep phenotypic profiling of 3D organotypic liver models
Biotechnology Journal ( IF 4.7 ) Pub Date : 2024-03-21 , DOI: 10.1002/biot.202300684 Stefania Koutsilieri 1 , Evgeniya Mickols 1, 2 , Ákos Végvári 3 , Volker M. Lauschke 1, 4, 5
Biotechnology Journal ( IF 4.7 ) Pub Date : 2024-03-21 , DOI: 10.1002/biot.202300684 Stefania Koutsilieri 1 , Evgeniya Mickols 1, 2 , Ákos Végvári 3 , Volker M. Lauschke 1, 4, 5
Affiliation
|
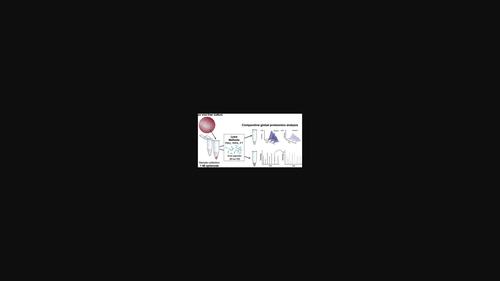
Organotypic human tissue models constitute promising systems to facilitate drug discovery and development. They allow to maintain native cellular phenotypes and functions, which enables long‐term pharmacokinetic and toxicity studies, as well as phenotypic screening. To trace relevant phenotypic changes back to specific targets or signaling pathways, comprehensive proteomic profiling is the gold‐standard. A multitude of proteomic workflows have been applied on 3D tissue models to quantify their molecular phenotypes; however, their impact on analytical results and biological conclusions in this context has not been evaluated. The performance of twelve mass spectrometry‐based global proteomic workflows that differed in the amount of cellular input, lysis protocols and quantification methods was compared for the analysis of primary human liver spheroids. Results differed majorly between protocols in the total number and subcellular compartment bias of identified proteins, which is particularly relevant for the reliable quantification of transporters and drug metabolizing enzymes. Using a model of metabolic dysfunction‐associated steatotic liver disease, we furthermore show that critical disease pathways are robustly identified using a standardized high throughput‐compatible workflow based on thermal lysis, even using only individual spheroids (1500 cells) as input. The results increase the applicability of proteomic profiling to phenotypic screens in organotypic microtissues and provide a scalable platform for deep phenotyping from limited biological material.
中文翻译:

用于 3D 器官型肝脏模型深度表型分析的蛋白质组工作流程
器官型人体组织模型构成了促进药物发现和开发的有前景的系统。它们可以维持天然细胞表型和功能,从而能够进行长期药代动力学和毒性研究以及表型筛选。为了追踪相关表型变化到特定靶点或信号通路,全面的蛋白质组分析是黄金标准。多种蛋白质组学工作流程已应用于 3D 组织模型,以量化其分子表型;然而,它们对这方面的分析结果和生物学结论的影响尚未得到评估。比较了 12 种基于质谱的全局蛋白质组工作流程的性能,这些工作流程在细胞输入量、裂解方案和定量方法方面有所不同,用于分析原代人肝球体。不同方案之间的结果差异主要在于已识别蛋白质的总数和亚细胞区室偏差,这对于转运蛋白和药物代谢酶的可靠定量特别相关。使用代谢功能障碍相关的脂肪肝病模型,我们进一步表明,使用基于热裂解的标准化高通量兼容工作流程,甚至仅使用单个球体(1500 个细胞)作为输入,可以稳健地识别关键疾病途径。这些结果增加了蛋白质组分析在器官型微组织表型筛选中的适用性,并为有限生物材料的深度表型分析提供了一个可扩展的平台。
更新日期:2024-03-21
中文翻译:

用于 3D 器官型肝脏模型深度表型分析的蛋白质组工作流程
器官型人体组织模型构成了促进药物发现和开发的有前景的系统。它们可以维持天然细胞表型和功能,从而能够进行长期药代动力学和毒性研究以及表型筛选。为了追踪相关表型变化到特定靶点或信号通路,全面的蛋白质组分析是黄金标准。多种蛋白质组学工作流程已应用于 3D 组织模型,以量化其分子表型;然而,它们对这方面的分析结果和生物学结论的影响尚未得到评估。比较了 12 种基于质谱的全局蛋白质组工作流程的性能,这些工作流程在细胞输入量、裂解方案和定量方法方面有所不同,用于分析原代人肝球体。不同方案之间的结果差异主要在于已识别蛋白质的总数和亚细胞区室偏差,这对于转运蛋白和药物代谢酶的可靠定量特别相关。使用代谢功能障碍相关的脂肪肝病模型,我们进一步表明,使用基于热裂解的标准化高通量兼容工作流程,甚至仅使用单个球体(1500 个细胞)作为输入,可以稳健地识别关键疾病途径。这些结果增加了蛋白质组分析在器官型微组织表型筛选中的适用性,并为有限生物材料的深度表型分析提供了一个可扩展的平台。



























 京公网安备 11010802027423号
京公网安备 11010802027423号