当前位置:
X-MOL 学术
›
Environ. Microbiol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Employing a triple metabarcoding approach to differentiate active, dormant and dead microeukaryotes in sediments
Environmental Microbiology ( IF 5.1 ) Pub Date : 2024-03-19 , DOI: 10.1111/1462-2920.16615 Huiwen Deng 1, 2 , Cui He 1 , Alexandra Z. Worden 3, 4 , Jun Gong 1, 2
Environmental Microbiology ( IF 5.1 ) Pub Date : 2024-03-19 , DOI: 10.1111/1462-2920.16615 Huiwen Deng 1, 2 , Cui He 1 , Alexandra Z. Worden 3, 4 , Jun Gong 1, 2
Affiliation
|
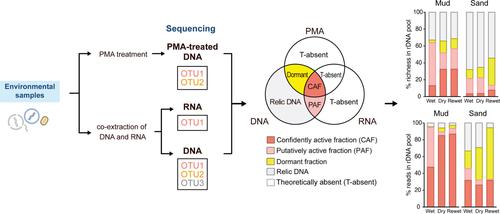
Microbial communities are commonly characterised through the metabarcoding of environmental DNA. This DNA originates from both viable (including dormant and active) and dead organisms, leading to recent efforts to distinguish between these states. In this study, we further these approaches by distinguishing not only between viable and dead cells but also between dormant and actively growing cells. This is achieved by sequencing both rRNA and rDNA, in conjunction with propidium monoazide cross‐linked rDNA, to partition the active, dormant and relic fractions in environmental samples. We apply this method to characterise the diversity and assemblage structure of these fractions of microeukaryotes in intertidal sediments during a wet‐dry‐rewet incubation cycle. Our findings indicate that a significant proportion of microeukaryotic phylotypes detected in the total rDNA pools originate from dormant and relic microeukaryotes in the sediments, both in terms of richness (dormant, 13 ± 2%; relic, 47 ± 5%) and read abundance (dormant, 20 ± 7%; relic, 14 ± 5%). The richness and sequence proportion of dormant microeukaryotes notably increase during the transition from wet to dry conditions. Statistical analyses suggest that the dynamics of diversity and assemblage structure across different activity fractions are influenced by various environmental drivers. Our strategy offers a versatile approach that can be adapted to characterise other microbes in a wide range of environments.
中文翻译:

采用三重元条形码方法区分沉积物中活跃、休眠和死亡的微真核生物
微生物群落通常通过环境 DNA 的元条形码来表征。这种 DNA 来源于存活的(包括休眠的和活跃的)和死亡的生物体,导致最近努力区分这些状态。在这项研究中,我们不仅区分活细胞和死亡细胞,还区分休眠细胞和活跃生长的细胞,进一步推进这些方法。这是通过对 rRNA 和 rDNA 进行测序,并结合单叠氮化丙啶交联的 rDNA,来区分环境样品中的活性、休眠和残余部分来实现的。我们应用这种方法来表征湿-干-再湿孵化周期期间潮间带沉积物中这些微真核生物部分的多样性和组合结构。我们的研究结果表明,在总 rDNA 库中检测到的微真核生物系统型的很大一部分源自沉积物中的休眠和遗迹微真核生物,无论是在丰富度(休眠,13 ± 2%;遗迹,47 ± 5%)还是读数丰度(休眠,20 ± 7%;遗迹,14 ± 5%)。休眠微真核生物的丰富度和序列比例在从潮湿到干燥条件的转变过程中显着增加。统计分析表明,不同活动部分的多样性和组合结构的动态受到各种环境驱动因素的影响。我们的策略提供了一种多功能方法,可用于表征各种环境中的其他微生物。
更新日期:2024-03-19
中文翻译:

采用三重元条形码方法区分沉积物中活跃、休眠和死亡的微真核生物
微生物群落通常通过环境 DNA 的元条形码来表征。这种 DNA 来源于存活的(包括休眠的和活跃的)和死亡的生物体,导致最近努力区分这些状态。在这项研究中,我们不仅区分活细胞和死亡细胞,还区分休眠细胞和活跃生长的细胞,进一步推进这些方法。这是通过对 rRNA 和 rDNA 进行测序,并结合单叠氮化丙啶交联的 rDNA,来区分环境样品中的活性、休眠和残余部分来实现的。我们应用这种方法来表征湿-干-再湿孵化周期期间潮间带沉积物中这些微真核生物部分的多样性和组合结构。我们的研究结果表明,在总 rDNA 库中检测到的微真核生物系统型的很大一部分源自沉积物中的休眠和遗迹微真核生物,无论是在丰富度(休眠,13 ± 2%;遗迹,47 ± 5%)还是读数丰度(休眠,20 ± 7%;遗迹,14 ± 5%)。休眠微真核生物的丰富度和序列比例在从潮湿到干燥条件的转变过程中显着增加。统计分析表明,不同活动部分的多样性和组合结构的动态受到各种环境驱动因素的影响。我们的策略提供了一种多功能方法,可用于表征各种环境中的其他微生物。



























 京公网安备 11010802027423号
京公网安备 11010802027423号