当前位置:
X-MOL 学术
›
Ecol. Lett.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
recolorize: An R package for flexible colour segmentation of biological images
Ecology Letters ( IF 8.8 ) Pub Date : 2024-02-05 , DOI: 10.1111/ele.14378 Hannah I. Weller 1, 2 , Anna E. Hiller 3 , Nathan P. Lord 4 , Steven M. Van Belleghem 5
Ecology Letters ( IF 8.8 ) Pub Date : 2024-02-05 , DOI: 10.1111/ele.14378 Hannah I. Weller 1, 2 , Anna E. Hiller 3 , Nathan P. Lord 4 , Steven M. Van Belleghem 5
Affiliation
|
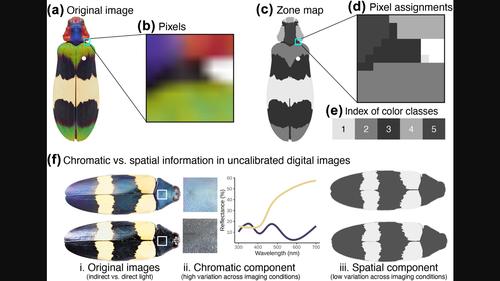
Colour pattern variation provides biological information in fields ranging from disease ecology to speciation dynamics. Comparing colour pattern geometries across images requires colour segmentation, where pixels in an image are assigned to one of a set of colour classes shared by all images. Manual methods for colour segmentation are slow and subjective, while automated methods can struggle with high technical variation in aggregate image sets. We present recolorize, an R package toolbox for human-subjective colour segmentation with functions for batch-processing low-variation image sets and additional tools for handling images from diverse (high-variation) sources. The package also includes export options for a variety of formats and colour analysis packages. This paper illustrates recolorize for three example datasets, including high variation, batch processing and combining with reflectance spectra, and demonstrates the downstream use of methods that rely on this output.
中文翻译:

recolorize:用于生物图像灵活颜色分割的 R 包
颜色模式变化提供了从疾病生态学到物种动态等领域的生物信息。比较图像之间的颜色图案几何形状需要颜色分割,其中图像中的像素被分配给所有图像共享的一组颜色类中的一个。颜色分割的手动方法缓慢且主观,而自动化方法则可能难以应对聚合图像集中的高技术变化。我们推出了 recolorize,这是一个用于人类主观颜色分割的 R 包工具箱,具有批处理低变化图像集的功能以及用于处理来自不同(高变化)来源的图像的附加工具。该软件包还包括各种格式的导出选项和颜色分析软件包。本文说明了三个示例数据集的重新着色,包括高变化、批处理和与反射光谱的结合,并演示了依赖此输出的方法的下游使用。
更新日期:2024-02-06
中文翻译:

recolorize:用于生物图像灵活颜色分割的 R 包
颜色模式变化提供了从疾病生态学到物种动态等领域的生物信息。比较图像之间的颜色图案几何形状需要颜色分割,其中图像中的像素被分配给所有图像共享的一组颜色类中的一个。颜色分割的手动方法缓慢且主观,而自动化方法则可能难以应对聚合图像集中的高技术变化。我们推出了 recolorize,这是一个用于人类主观颜色分割的 R 包工具箱,具有批处理低变化图像集的功能以及用于处理来自不同(高变化)来源的图像的附加工具。该软件包还包括各种格式的导出选项和颜色分析软件包。本文说明了三个示例数据集的重新着色,包括高变化、批处理和与反射光谱的结合,并演示了依赖此输出的方法的下游使用。



























 京公网安备 11010802027423号
京公网安备 11010802027423号