当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Gentle Introduction to the Statistical Foundations of False Discovery Rate in Quantitative Proteomics
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2017-11-14 00:00:00 , DOI: 10.1021/acs.jproteome.7b00170 Thomas Burger 1
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2017-11-14 00:00:00 , DOI: 10.1021/acs.jproteome.7b00170 Thomas Burger 1
Affiliation
|
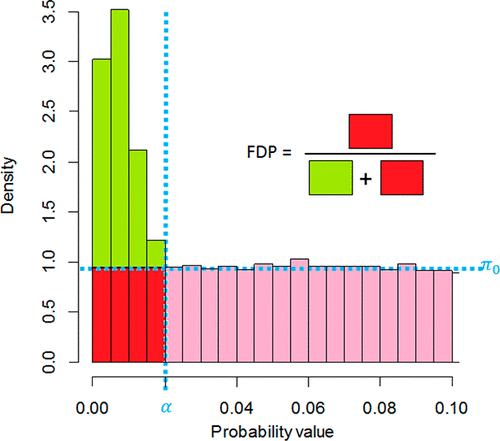
The vocabulary of theoretical statistics can be difficult to embrace from the viewpoint of computational proteomics research, even though the notions it conveys are essential to publication guidelines. For example, “adjusted p-values”, “q-values”, and “false discovery rates” are essentially similar concepts, whereas “false discovery rate” and “false discovery proportion” must not be confused, even though “rate” and “proportion” are related in everyday language. In the interdisciplinary context of proteomics, such subtleties may cause misunderstandings. This article aims to provide an easy-to-understand explanation of these four notions (and a few other related ones). Their statistical foundations are dealt with from a perspective that largely relies on intuition, addressing mainly protein quantification but also, to some extent, peptide identification. In addition, a clear distinction is made between concepts that define an individual property (i.e., related to a peptide or a protein) and those that define a set property (i.e., related to a list of peptides or proteins).
中文翻译:

定量蛋白质组学中错误发现率统计基础的简要介绍
从计算蛋白质组学研究的角度来看,理论统计学的词汇可能难以理解,即使它传达的概念对于出版指南至关重要。例如,“调整后的p值”,“ q-值”和“错误发现率”在本质上是相似的概念,尽管“速率”和“比例”在日常语言中是相关的,但也不能混淆“错误发现率”和“错误发现率”。在蛋白质组学的跨学科背景下,这种微妙之处可能会引起误解。本文旨在提供对这四个概念(以及其他一些相关概念)的易于理解的解释。他们的统计基础从很大程度上依赖于直觉的角度来处理,主要解决蛋白质定量问题,而且在某种程度上还解决了肽段鉴定问题。此外,在定义单个属性(即与肽或蛋白质有关)的概念与定义集合属性(即与肽或蛋白质列表有关)的概念之间有明显的区别。
更新日期:2017-11-15
中文翻译:

定量蛋白质组学中错误发现率统计基础的简要介绍
从计算蛋白质组学研究的角度来看,理论统计学的词汇可能难以理解,即使它传达的概念对于出版指南至关重要。例如,“调整后的p值”,“ q-值”和“错误发现率”在本质上是相似的概念,尽管“速率”和“比例”在日常语言中是相关的,但也不能混淆“错误发现率”和“错误发现率”。在蛋白质组学的跨学科背景下,这种微妙之处可能会引起误解。本文旨在提供对这四个概念(以及其他一些相关概念)的易于理解的解释。他们的统计基础从很大程度上依赖于直觉的角度来处理,主要解决蛋白质定量问题,而且在某种程度上还解决了肽段鉴定问题。此外,在定义单个属性(即与肽或蛋白质有关)的概念与定义集合属性(即与肽或蛋白质列表有关)的概念之间有明显的区别。











































 京公网安备 11010802027423号
京公网安备 11010802027423号