当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Improved Prediction of Bovine Leucocyte Antigens (BoLA) Presented Ligands by Use of Mass-Spectrometry-Determined Ligand and in Vitro Binding Data
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2017-11-14 00:00:00 , DOI: 10.1021/acs.jproteome.7b00675 Morten Nielsen 1, 2 , Tim Connelley 3 , Nicola Ternette 4
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2017-11-14 00:00:00 , DOI: 10.1021/acs.jproteome.7b00675 Morten Nielsen 1, 2 , Tim Connelley 3 , Nicola Ternette 4
Affiliation
|
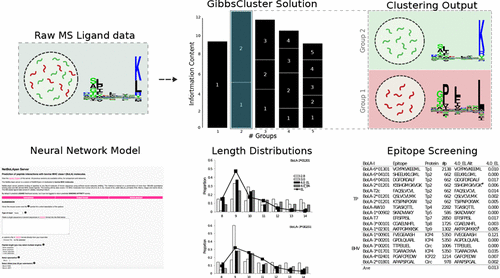
Peptide binding to MHC class I molecules is the single most selective step in antigen presentation and the strongest single correlate to peptide cellular immunogenicity. The cost of experimentally characterizing the rules of peptide presentation for a given MHC-I molecule is extensive, and predictors of peptide–MHC interactions constitute an attractive alternative. Recently, an increasing amount of MHC presented peptides identified by mass spectrometry (MS ligands) has been published. Handling and interpretation of MS ligand data is, in general, challenging due to the polyspecificity nature of the data. We here outline a general pipeline for dealing with this challenge and accurately annotate ligands to the relevant MHC-I molecule they were eluted from by use of GibbsClustering and binding motif information inferred from in silico models. We illustrate the approach here in the context of MHC-I molecules (BoLA) of cattle. Next, we demonstrate how such annotated BoLA MS ligand data can readily be integrated with in vitro binding affinity data in a prediction model with very high and unprecedented performance for identification of BoLA-I restricted T-cell epitopes. The prediction model is freely available at http://www.cbs.dtu.dk/services/NetMHCpan/NetBoLApan. The approach has here been applied to the BoLA-I system, but the pipeline is readily applicable to MHC systems in other species.
中文翻译:

使用质谱测定的配体和体外结合数据改进对牛白细胞抗原(BoLA)呈递配体的预测
肽与MHC I类分子的结合是抗原呈递中最选择性的步骤,并且最强的一个与肽细胞免疫原性相关。对于给定的MHC-1分子,实验表征肽呈递规则的成本是巨大的,而肽与MHC相互作用的预测因子则是一个有吸引力的选择。最近,已经公开了通过质谱法(MS配体)鉴定的MHC呈递肽的数量增加。由于数据的多特异性性质,MS配体数据的处理和解释通常具有挑战性。我们在这里概述了应对这一挑战的一般流程,并通过使用GibbsClustering和结合计算机模型推断的基序信息,准确地注释了配体到相关MHC-1分子的配体。我们在牛的MHC-1分子(BoLA)的背景下说明了此方法。接下来,我们展示了如何将这种带注释的BoLA MS配体数据与体外结合亲和力数据轻松集成到预测模型中,该模型具有很高的空前性能,可用于鉴定BoLA-1限制性T细胞表位。该预测模型可在以下位置免费获得http://www.cbs.dtu.dk/services/NetMHCpan/NetBoLApan。该方法已在此处应用于BoLA-I系统,但该管道很容易适用于其他物种的MHC系统。
更新日期:2017-11-15
中文翻译:

使用质谱测定的配体和体外结合数据改进对牛白细胞抗原(BoLA)呈递配体的预测
肽与MHC I类分子的结合是抗原呈递中最选择性的步骤,并且最强的一个与肽细胞免疫原性相关。对于给定的MHC-1分子,实验表征肽呈递规则的成本是巨大的,而肽与MHC相互作用的预测因子则是一个有吸引力的选择。最近,已经公开了通过质谱法(MS配体)鉴定的MHC呈递肽的数量增加。由于数据的多特异性性质,MS配体数据的处理和解释通常具有挑战性。我们在这里概述了应对这一挑战的一般流程,并通过使用GibbsClustering和结合计算机模型推断的基序信息,准确地注释了配体到相关MHC-1分子的配体。我们在牛的MHC-1分子(BoLA)的背景下说明了此方法。接下来,我们展示了如何将这种带注释的BoLA MS配体数据与体外结合亲和力数据轻松集成到预测模型中,该模型具有很高的空前性能,可用于鉴定BoLA-1限制性T细胞表位。该预测模型可在以下位置免费获得http://www.cbs.dtu.dk/services/NetMHCpan/NetBoLApan。该方法已在此处应用于BoLA-I系统,但该管道很容易适用于其他物种的MHC系统。











































 京公网安备 11010802027423号
京公网安备 11010802027423号