当前位置:
X-MOL 学术
›
Mol. Pharmaceutics
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Comparison of Deep Learning With Multiple Machine Learning Methods and Metrics Using Diverse Drug Discovery Data Sets
Molecular Pharmaceutics ( IF 4.9 ) Pub Date : 2017-11-13 00:00:00 , DOI: 10.1021/acs.molpharmaceut.7b00578 Alexandru Korotcov 1 , Valery Tkachenko 1 , Daniel P. Russo 2, 3 , Sean Ekins 2
Molecular Pharmaceutics ( IF 4.9 ) Pub Date : 2017-11-13 00:00:00 , DOI: 10.1021/acs.molpharmaceut.7b00578 Alexandru Korotcov 1 , Valery Tkachenko 1 , Daniel P. Russo 2, 3 , Sean Ekins 2
Affiliation
|
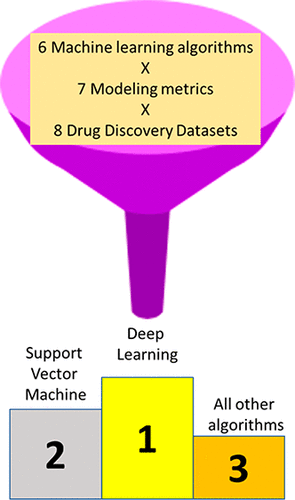
Machine learning methods have been applied to many data sets in pharmaceutical research for several decades. The relative ease and availability of fingerprint type molecular descriptors paired with Bayesian methods resulted in the widespread use of this approach for a diverse array of end points relevant to drug discovery. Deep learning is the latest machine learning algorithm attracting attention for many of pharmaceutical applications from docking to virtual screening. Deep learning is based on an artificial neural network with multiple hidden layers and has found considerable traction for many artificial intelligence applications. We have previously suggested the need for a comparison of different machine learning methods with deep learning across an array of varying data sets that is applicable to pharmaceutical research. End points relevant to pharmaceutical research include absorption, distribution, metabolism, excretion, and toxicity (ADME/Tox) properties, as well as activity against pathogens and drug discovery data sets. In this study, we have used data sets for solubility, probe-likeness, hERG, KCNQ1, bubonic plague, Chagas, tuberculosis, and malaria to compare different machine learning methods using FCFP6 fingerprints. These data sets represent whole cell screens, individual proteins, physicochemical properties as well as a data set with a complex end point. Our aim was to assess whether deep learning offered any improvement in testing when assessed using an array of metrics including AUC, F1 score, Cohen’s kappa, Matthews correlation coefficient and others. Based on ranked normalized scores for the metrics or data sets Deep Neural Networks (DNN) ranked higher than SVM, which in turn was ranked higher than all the other machine learning methods. Visualizing these properties for training and test sets using radar type plots indicates when models are inferior or perhaps over trained. These results also suggest the need for assessing deep learning further using multiple metrics with much larger scale comparisons, prospective testing as well as assessment of different fingerprints and DNN architectures beyond those used.
中文翻译:

使用多种药物发现数据集将深度学习与多种机器学习方法和指标进行比较
数十年来,机器学习方法已应用于药物研究中的许多数据集。与贝叶斯方法配对的指纹类型分子描述符的相对易用性和可用性导致该方法被广泛用于与药物发现有关的各种终点。深度学习是最新的机器学习算法,从对接到虚拟筛选,在许多制药应用中都引起了人们的关注。深度学习基于具有多个隐藏层的人工神经网络,并已为许多人工智能应用找到了可观的吸引力。我们以前曾建议需要将适用于药物研究的各种机器学习方法与深度学习跨一系列可变数据集进行比较。与药物研究相关的终点包括吸收,分布,代谢,排泄和毒性(ADME / Tox)特性,以及针对病原体和药物发现数据集的活性。在这项研究中,我们使用了溶解度,探针样,hERG,KCNQ1,布氏鼠疫,恰加斯,结核和疟疾的数据集,以比较使用FCFP6指纹的不同机器学习方法。这些数据集代表整个细胞的筛选,单个蛋白质,理化特性以及具有复杂终点的数据集。我们的目标是评估使用一系列指标(包括AUC,F1得分,科恩卡伯值,马修斯相关系数等)进行评估时,深度学习是否对测试有所改善。根据指标或数据集的归一化分数,深度神经网络(DNN)的排名高于SVM,而SVM的排名则高于所有其他机器学习方法。使用雷达类型图将这些属性可视化以用于训练和测试集,可指示何时模型较差或训练过度。这些结果还表明,需要使用具有更大比例比较的多个指标,前瞻性测试以及除使用的指纹和DNN架构之外的其他指纹和DNN架构来进一步评估深度学习。
更新日期:2017-11-13
中文翻译:

使用多种药物发现数据集将深度学习与多种机器学习方法和指标进行比较
数十年来,机器学习方法已应用于药物研究中的许多数据集。与贝叶斯方法配对的指纹类型分子描述符的相对易用性和可用性导致该方法被广泛用于与药物发现有关的各种终点。深度学习是最新的机器学习算法,从对接到虚拟筛选,在许多制药应用中都引起了人们的关注。深度学习基于具有多个隐藏层的人工神经网络,并已为许多人工智能应用找到了可观的吸引力。我们以前曾建议需要将适用于药物研究的各种机器学习方法与深度学习跨一系列可变数据集进行比较。与药物研究相关的终点包括吸收,分布,代谢,排泄和毒性(ADME / Tox)特性,以及针对病原体和药物发现数据集的活性。在这项研究中,我们使用了溶解度,探针样,hERG,KCNQ1,布氏鼠疫,恰加斯,结核和疟疾的数据集,以比较使用FCFP6指纹的不同机器学习方法。这些数据集代表整个细胞的筛选,单个蛋白质,理化特性以及具有复杂终点的数据集。我们的目标是评估使用一系列指标(包括AUC,F1得分,科恩卡伯值,马修斯相关系数等)进行评估时,深度学习是否对测试有所改善。根据指标或数据集的归一化分数,深度神经网络(DNN)的排名高于SVM,而SVM的排名则高于所有其他机器学习方法。使用雷达类型图将这些属性可视化以用于训练和测试集,可指示何时模型较差或训练过度。这些结果还表明,需要使用具有更大比例比较的多个指标,前瞻性测试以及除使用的指纹和DNN架构之外的其他指纹和DNN架构来进一步评估深度学习。



























 京公网安备 11010802027423号
京公网安备 11010802027423号