当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
PhoStar: Identifying Tandem Mass Spectra of Phosphorylated Peptides before Database Search
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2017-11-02 00:00:00 , DOI: 10.1021/acs.jproteome.7b00563 Sebastian Dorl 1 , Stephan Winkler 1 , Karl Mechtler 2, 3 , Viktoria Dorfer 1
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2017-11-02 00:00:00 , DOI: 10.1021/acs.jproteome.7b00563 Sebastian Dorl 1 , Stephan Winkler 1 , Karl Mechtler 2, 3 , Viktoria Dorfer 1
Affiliation
|
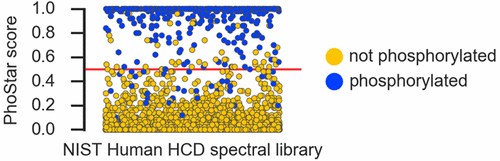
Standard proteomics workflows use tandem mass spectrometry followed by sequence database search to analyze complex biological samples. The identification of proteins carrying post-translational modifications, for example, phosphorylation, is typically addressed by allowing variable modifications in the searched sequences. Accounting for these variations exponentially increases the combinatorial space in the database, which leads to increased processing times and more false positive identifications. The here-presented tool PhoStar identifies spectra that originate from phosphorylated peptides before database search using a supervised machine learning approach. The model for the prediction of phosphorylation was trained and validated with an accuracy of 97.6% on a large set of high-confidence spectra collected from publicly available experimental data. Its power was further validated by predicting phosphorylation in the complete NIST human and mouse high collision-dissociation spectral libraries, achieving an accuracy of 98.2 and 97.9%, respectively. We demonstrate the application of PhoStar by using it for spectra filtering before database search. In database search of HeLa samples the peptide search space was reduced by 27–66% while finding at least 97% of total peptide identifications (at 1% FDR) compared with a standard workflow.
中文翻译:

PhoStar:在数据库搜索之前识别磷酸化肽的串联质谱
标准蛋白质组学工作流程使用串联质谱法,然后使用序列数据库搜索来分析复杂的生物样品。携带翻译后修饰(例如磷酸化)的蛋白质的鉴定通常通过在搜索到的序列中进行可变修饰来解决。考虑这些变化会成倍增加数据库中的组合空间,从而导致处理时间增加和错误的肯定标识。本文介绍的工具PhoStar使用有监督的机器学习方法在数据库搜索之前识别源自磷酸化肽段的光谱。预测并预测了磷酸化的模型,准确性为97。从公开的实验数据中收集的大量高可信度光谱中,只有6%。通过预测完整的NIST人类和小鼠高碰撞解离光谱库中的磷酸化,可以进一步验证其功能,分别达到98.2%和97.9%的准确度。我们通过在数据库搜索之前使用PhoStar进行光谱过滤来演示PhoStar的应用。与标准工作流程相比,在HeLa样品的数据库搜索中,肽搜索空间减少了27–66%,同时发现了至少97%的总肽鉴定(FDR为1%)。我们通过在数据库搜索之前使用PhoStar进行光谱过滤来演示PhoStar的应用。与标准工作流程相比,在HeLa样品的数据库搜索中,肽搜索空间减少了27–66%,同时发现了至少97%的总肽鉴定(FDR为1%)。我们通过在数据库搜索之前使用PhoStar进行光谱过滤来演示PhoStar的应用。与标准工作流程相比,在HeLa样品的数据库搜索中,肽搜索空间减少了27–66%,同时发现了至少97%的总肽鉴定(FDR为1%)。
更新日期:2017-11-03
中文翻译:

PhoStar:在数据库搜索之前识别磷酸化肽的串联质谱
标准蛋白质组学工作流程使用串联质谱法,然后使用序列数据库搜索来分析复杂的生物样品。携带翻译后修饰(例如磷酸化)的蛋白质的鉴定通常通过在搜索到的序列中进行可变修饰来解决。考虑这些变化会成倍增加数据库中的组合空间,从而导致处理时间增加和错误的肯定标识。本文介绍的工具PhoStar使用有监督的机器学习方法在数据库搜索之前识别源自磷酸化肽段的光谱。预测并预测了磷酸化的模型,准确性为97。从公开的实验数据中收集的大量高可信度光谱中,只有6%。通过预测完整的NIST人类和小鼠高碰撞解离光谱库中的磷酸化,可以进一步验证其功能,分别达到98.2%和97.9%的准确度。我们通过在数据库搜索之前使用PhoStar进行光谱过滤来演示PhoStar的应用。与标准工作流程相比,在HeLa样品的数据库搜索中,肽搜索空间减少了27–66%,同时发现了至少97%的总肽鉴定(FDR为1%)。我们通过在数据库搜索之前使用PhoStar进行光谱过滤来演示PhoStar的应用。与标准工作流程相比,在HeLa样品的数据库搜索中,肽搜索空间减少了27–66%,同时发现了至少97%的总肽鉴定(FDR为1%)。我们通过在数据库搜索之前使用PhoStar进行光谱过滤来演示PhoStar的应用。与标准工作流程相比,在HeLa样品的数据库搜索中,肽搜索空间减少了27–66%,同时发现了至少97%的总肽鉴定(FDR为1%)。











































 京公网安备 11010802027423号
京公网安备 11010802027423号