当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Identification and Validation of Human Missing Proteins and Peptides in Public Proteome Databases: Data Mining Strategy
Journal of Proteome Research ( IF 4.4 ) Pub Date : 2017-10-31 00:00:00 , DOI: 10.1021/acs.jproteome.7b00423 Amr Elguoshy 1, 2, 3 , Yoshitoshi Hirao 1 , Bo Xu 1 , Suguru Saito 1 , Ali F. Quadery 1 , Keiko Yamamoto 1 , Toshiaki Mitsui 2 , Tadashi Yamamoto 1 ,
Journal of Proteome Research ( IF 4.4 ) Pub Date : 2017-10-31 00:00:00 , DOI: 10.1021/acs.jproteome.7b00423 Amr Elguoshy 1, 2, 3 , Yoshitoshi Hirao 1 , Bo Xu 1 , Suguru Saito 1 , Ali F. Quadery 1 , Keiko Yamamoto 1 , Toshiaki Mitsui 2 , Tadashi Yamamoto 1 ,
Affiliation
|
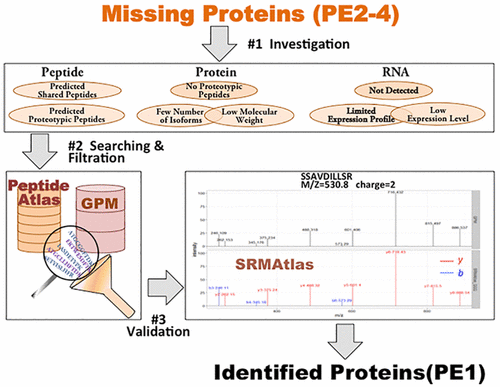
In an attempt to complete human proteome project (HPP), Chromosome-Centric Human Proteome Project (C-HPP) launched the journey of missing protein (MP) investigation in 2012. However, 2579 and 572 protein entries in the neXtProt (2017-1) are still considered as missing and uncertain proteins, respectively. Thus, in this study, we proposed a pipeline to analyze, identify, and validate human missing and uncertain proteins in open-access transcriptomics and proteomics databases. Analysis of RNA expression pattern for missing proteins in Human protein Atlas showed that 28% of them, such as Olfactory receptor 1I1 (O60431), had no RNA expression, suggesting the necessity to consider uncommon tissues for transcriptomic and proteomic studies. Interestingly, 21% had elevated expression level in a particular tissue (tissue-enriched proteins), indicating the importance of targeting such proteins in their elevated tissues. Additionally, the analysis of RNA expression level for missing proteins showed that 95% had no or low expression level (0–10 transcripts per million), indicating that low abundance is one of the major obstacles facing the detection of missing proteins. Moreover, missing proteins are predicted to generate fewer predicted unique tryptic peptides than the identified proteins. Searching for these predicted unique tryptic peptides that correspond to missing and uncertain proteins in the experimental peptide list of open-access MS-based databases (PA, GPM) resulted in the detection of 402 missing and 19 uncertain proteins with at least two unique peptides (≥9 aa) at <(5 × 10–4)% FDR. Finally, matching the native spectra for the experimentally detected peptides with their SRMAtlas synthetic counterparts at three transition sources (QQQ, QTOF, QTRAP) gave us an opportunity to validate 41 missing proteins by ≥2 proteotypic peptides.
中文翻译:

蛋白质组数据库中人类缺失蛋白和多肽的鉴定和验证:数据挖掘策略
为了完成人类蛋白质组计划(HPP),2012年染色体中心人类蛋白质组计划(C-HPP)启动了缺失蛋白质(MP)研究的旅程。但是,neXtProt(2017-1)中有2579和572个蛋白质条目)仍分别被认为是蛋白质缺失和不确定。因此,在这项研究中,我们提出了一条管道,用于分析,识别和验证开放获取转录组学和蛋白质组学数据库中的人类缺失和不确定的蛋白质。分析人类蛋白质图谱中缺失蛋白质的RNA表达模式后发现,其中28%如嗅觉受体1I1(O60431),没有RNA表达,这表明有必要考虑进行转录组学和蛋白质组学研究的不常见组织。有趣的是,有21%的蛋白质在特定组织(富含组织的蛋白质)中的表达水平升高,表明在此类蛋白质的升高的组织中靶向此类蛋白质的重要性。此外,对缺失蛋白的RNA表达水平的分析表明,有95%的蛋白没有表达水平或表达水平很低(百万分之0-10转录本),这表明低丰度是检测缺失蛋白的主要障碍之一。此外,与鉴定的蛋白质相比,预测缺失的蛋白质产生的预测的独特胰蛋白酶消化的肽更少。在基于开放获取的基于MS的数据库(PA,–4)%FDR。最后,在三个转换源(QQQ,QTOF,QTRAP)上将实验检测到的肽段的天然光谱与其SRMAtlas合成对应物进行匹配,使我们有机会通过≥2个蛋白型肽段验证41种缺失蛋白。
更新日期:2017-11-01
中文翻译:

蛋白质组数据库中人类缺失蛋白和多肽的鉴定和验证:数据挖掘策略
为了完成人类蛋白质组计划(HPP),2012年染色体中心人类蛋白质组计划(C-HPP)启动了缺失蛋白质(MP)研究的旅程。但是,neXtProt(2017-1)中有2579和572个蛋白质条目)仍分别被认为是蛋白质缺失和不确定。因此,在这项研究中,我们提出了一条管道,用于分析,识别和验证开放获取转录组学和蛋白质组学数据库中的人类缺失和不确定的蛋白质。分析人类蛋白质图谱中缺失蛋白质的RNA表达模式后发现,其中28%如嗅觉受体1I1(O60431),没有RNA表达,这表明有必要考虑进行转录组学和蛋白质组学研究的不常见组织。有趣的是,有21%的蛋白质在特定组织(富含组织的蛋白质)中的表达水平升高,表明在此类蛋白质的升高的组织中靶向此类蛋白质的重要性。此外,对缺失蛋白的RNA表达水平的分析表明,有95%的蛋白没有表达水平或表达水平很低(百万分之0-10转录本),这表明低丰度是检测缺失蛋白的主要障碍之一。此外,与鉴定的蛋白质相比,预测缺失的蛋白质产生的预测的独特胰蛋白酶消化的肽更少。在基于开放获取的基于MS的数据库(PA,–4)%FDR。最后,在三个转换源(QQQ,QTOF,QTRAP)上将实验检测到的肽段的天然光谱与其SRMAtlas合成对应物进行匹配,使我们有机会通过≥2个蛋白型肽段验证41种缺失蛋白。



























 京公网安备 11010802027423号
京公网安备 11010802027423号