Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
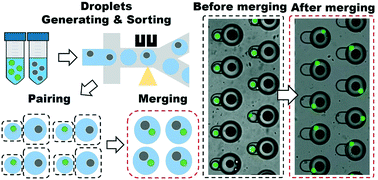
Deterministic droplet-based co-encapsulation and pairing of microparticles via active sorting and downstream merging
Lab on a Chip ( IF 6.1 ) Pub Date : 2017-09-22 00:00:00 , DOI: 10.1039/c7lc00745k Meng Ting Chung 1, 2, 3, 4 , Daniel Núñez 2, 3, 4, 5 , Dawen Cai 2, 3, 4, 5, 6 , Katsuo Kurabayashi 1, 2, 3, 4, 7
Lab on a Chip ( IF 6.1 ) Pub Date : 2017-09-22 00:00:00 , DOI: 10.1039/c7lc00745k Meng Ting Chung 1, 2, 3, 4 , Daniel Núñez 2, 3, 4, 5 , Dawen Cai 2, 3, 4, 5, 6 , Katsuo Kurabayashi 1, 2, 3, 4, 7
Affiliation
|
Co-encapsulation of two distinct particles within microfluidic droplets provides the means to achieve various high-throughput single-cell assays, such as biochemical reactions and cell–cell interactions in small isolated volumes. However, limited by the Poisson statistics, the co-encapsulation rate of the conventional co-flow approach is low even under optimal conditions. Only up to 13.5% of droplets precisely contain a pair of two distinct particles, while the rest, either being empty or encapsulating unpaired particles become wastes. Thus, the low co-encapsulation efficiency makes droplet-based assays impractical in biological applications involving low abundant bioparticles. In this paper, we present a highly promising droplet merging strategy to increase the co-encapsulation efficiency. Our method first enriches droplets exactly encapsulating a single particle via fluorescence or scattering-light activated sorting. Then, two droplets, each with a distinct particle, are precisely one-to-one paired and merged in a novel microwell device. This deterministic approach overcomes the Poisson statistics limitation facing conventional stochastic methods, yielding an up to 90% post-sorting particle capture rate and an overall 88.1% co-encapsulation rate. With its superior single-particle pairing performance, our system provides a promising technological platform to enable highly efficient microdroplet assays.
更新日期:2017-10-26









































 京公网安备 11010802027423号
京公网安备 11010802027423号