PLOS Biology ( IF 7.8 ) Pub Date : 2017-10-19 , DOI: 10.1371/journal.pbio.2004045 Jean-Louis Plouhinec , Sofía Medina-Ruiz , Caroline Borday , Elsa Bernard , Jean-Philippe Vert , Michael B. Eisen , Richard M. Harland , Anne H. Monsoro-Burq
|
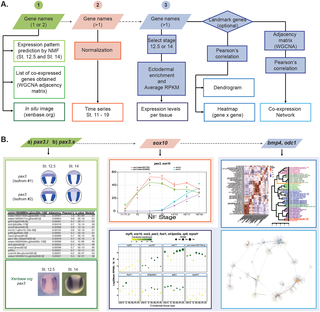
During vertebrate neurulation, the embryonic ectoderm is patterned into lineage progenitors for neural plate, neural crest, placodes and epidermis. Here, we use Xenopus laevis embryos to analyze the spatial and temporal transcriptome of distinct ectodermal domains in the course of neurulation, during the establishment of cell lineages. In order to define the transcriptome of small groups of cells from a single germ layer and to retain spatial information, dorsal and ventral ectoderm was subdivided along the anterior-posterior and medial-lateral axes by microdissections. Principal component analysis on the transcriptomes of these ectoderm fragments primarily identifies embryonic axes and temporal dynamics. This provides a genetic code to define positional information of any ectoderm sample along the anterior-posterior and dorsal-ventral axes directly from its transcriptome. In parallel, we use nonnegative matrix factorization to predict enhanced gene expression maps onto early and mid-neurula embryos, and specific signatures for each ectoderm area. The clustering of spatial and temporal datasets allowed detection of multiple biologically relevant groups (e.g., Wnt signaling, neural crest development, sensory placode specification, ciliogenesis, germ layer specification). We provide an interactive network interface, EctoMap, for exploring synexpression relationships among genes expressed in the neurula, and suggest several strategies to use this comprehensive dataset to address questions in developmental biology as well as stem cell or cancer research.
中文翻译:

外胚层发育的分子图谱定义了脊椎动物的神经,神经neural,斑块和非神经祖细胞身份
在脊椎动物的培养过程中,将胚外胚层图案化成谱系祖细胞,用于神经板,神经c,斑块和表皮。在这里,我们使用非洲爪蟾胚胎在细胞谱系建立过程中分析神经元在形成过程中不同外胚层结构域的时空转录组。为了定义来自单个胚层的小群细胞的转录组并保留空间信息,通过显微解剖将前后外胚轴细分。这些外胚层片段的转录组的主成分分析主要确定胚胎轴和时间动态。这提供了一个遗传密码,可直接从其转录组定义沿前后轴和后腹轴的任何外胚层样品的位置信息。同时,我们使用非负矩阵分解来预测早期和中期神经胚上增强的基因表达图,以及每个外胚层区域的特定签名。空间和时间数据集的聚类允许检测多个生物学相关的组(例如,Wnt信号,神经development发育,感觉斑规范,纤毛发生,胚层规范)。我们提供了一个交互式网络接口EctoMap,用于探索神经元中表达的基因之间的共表达关系,并提出几种策略来使用这一综合数据集来解决发育生物学以及干细胞或癌症研究中的问题。











































 京公网安备 11010802027423号
京公网安备 11010802027423号