当前位置:
X-MOL 学术
›
RSC Med. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A systematic analysis of atomic protein–ligand interactions in the PDB
RSC Medicinal Chemistry ( IF 4.1 ) Pub Date : 2017-09-26 00:00:00 , DOI: 10.1039/c7md00381a Renato Ferreira de Freitas 1, 2, 3, 4 , Matthieu Schapira 1, 2, 3, 4, 5
RSC Medicinal Chemistry ( IF 4.1 ) Pub Date : 2017-09-26 00:00:00 , DOI: 10.1039/c7md00381a Renato Ferreira de Freitas 1, 2, 3, 4 , Matthieu Schapira 1, 2, 3, 4, 5
Affiliation
|
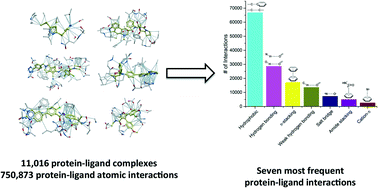
As the protein databank (PDB) recently passed the cap of 123 456 structures, it stands more than ever as an important resource not only to analyze structural features of specific biological systems, but also to study the prevalence of structural patterns observed in a large body of unrelated structures, that may reflect rules governing protein folding or molecular recognition. Here, we compiled a list of 11 016 unique structures of small-molecule ligands bound to proteins – 6444 of which have experimental binding affinity – representing 750 873 protein–ligand atomic interactions, and analyzed the frequency, geometry and impact of each interaction type. We find that hydrophobic interactions are generally enriched in high-efficiency ligands, but polar interactions are over-represented in fragment inhibitors. While most observations extracted from the PDB will be familiar to seasoned medicinal chemists, less expected findings, such as the high number of C–H⋯O hydrogen bonds or the relatively frequent amide–π stacking between the backbone amide of proteins and aromatic rings of ligands, uncover underused ligand design strategies.
中文翻译:

对PDB中原子蛋白质-配体相互作用的系统分析
由于蛋白质数据库(PDB)最近超过了123 456个结构的上限,因此它比以往任何时候都成为重要的资源,不仅可以分析特定生物系统的结构特征,而且可以研究在大人体中观察到的结构模式的普遍性不相关的结构,可能反映了控制蛋白质折叠或分子识别的规则。在这里,我们列出了与蛋白质结合的11 016个独特的小分子配体结构列表,其中6444个具有实验性结合亲和力,代表750 873个蛋白质与配体的原子相互作用,并分析了每种相互作用类型的频率,几何形状和影响。我们发现疏水性相互作用通常富含高效配体,但极性相互作用在片段抑制剂中却过分代表。
更新日期:2017-10-18
中文翻译:

对PDB中原子蛋白质-配体相互作用的系统分析
由于蛋白质数据库(PDB)最近超过了123 456个结构的上限,因此它比以往任何时候都成为重要的资源,不仅可以分析特定生物系统的结构特征,而且可以研究在大人体中观察到的结构模式的普遍性不相关的结构,可能反映了控制蛋白质折叠或分子识别的规则。在这里,我们列出了与蛋白质结合的11 016个独特的小分子配体结构列表,其中6444个具有实验性结合亲和力,代表750 873个蛋白质与配体的原子相互作用,并分析了每种相互作用类型的频率,几何形状和影响。我们发现疏水性相互作用通常富含高效配体,但极性相互作用在片段抑制剂中却过分代表。











































 京公网安备 11010802027423号
京公网安备 11010802027423号