当前位置:
X-MOL 学术
›
ACS Chem. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Observation by Real-Time NMR and Interpretation of Length- and Location-Dependent Deamination Activity of APOBEC3B
ACS Chemical Biology ( IF 3.5 ) Pub Date : 2017-10-03 00:00:00 , DOI: 10.1021/acschembio.7b00662 Li Wan 1 , Takashi Nagata 1 , Ryo Morishita 2 , Akifumi Takaori-Kondo 3 , Masato Katahira 1
ACS Chemical Biology ( IF 3.5 ) Pub Date : 2017-10-03 00:00:00 , DOI: 10.1021/acschembio.7b00662 Li Wan 1 , Takashi Nagata 1 , Ryo Morishita 2 , Akifumi Takaori-Kondo 3 , Masato Katahira 1
Affiliation
|
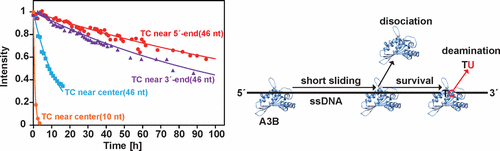
Human APOBEC3B (A3B) deaminates a cytosine into a uracil in single-stranded (ss) DNA, resulting in human cancers. A3B’s deamination activity is conferred by its C-terminal domain (CTD). However, little is known about the mechanism by which target sequences are searched and deaminated. Here, we applied a real-time NMR method to elucidate the deamination properties. We found that A3B CTD shows higher activity toward its target sequence in short ssDNA and efficiently deaminates a target sequence located near the center of ssDNA. These properties are quite different from those of well-studied APOBEC3G, which shows higher activity toward its target sequence in long ssDNA and one located close to the 5′-end. The unique properties of the A3B CTD can be rationally interpreted by considering that after nonspecific binding to ssDNA, A3B slides only for a relatively short distance and tends to dissociate from the ssDNA before reaching the target sequence.
中文翻译:

实时NMR观察和解释APOBEC3B的长度和位置相关的脱氨活性
人类APOBEC3B(A3B)将单链(ss)DNA中的胞嘧啶脱氨成尿嘧啶,从而导致人类癌症。A3B的脱氨活性由其C末端结构域(CTD)赋予。但是,关于靶序列被搜索和脱氨的机理知之甚少。在这里,我们应用了实时NMR方法来阐明脱氨性能。我们发现,A3B CTD在短的ssDNA中对其目标序列显示更高的活性,并有效地使位于ssDNA中心附近的目标序列脱氨。这些特性与经过充分研究的APOBEC3G完全不同,APOBEC3G在长ssDNA中显示出对其靶序列的更高活性,并且靠近5'端。考虑到在与ssDNA非特异性结合后,A3B CTD的独特性质可以得到合理的解释。
更新日期:2017-10-03
中文翻译:

实时NMR观察和解释APOBEC3B的长度和位置相关的脱氨活性
人类APOBEC3B(A3B)将单链(ss)DNA中的胞嘧啶脱氨成尿嘧啶,从而导致人类癌症。A3B的脱氨活性由其C末端结构域(CTD)赋予。但是,关于靶序列被搜索和脱氨的机理知之甚少。在这里,我们应用了实时NMR方法来阐明脱氨性能。我们发现,A3B CTD在短的ssDNA中对其目标序列显示更高的活性,并有效地使位于ssDNA中心附近的目标序列脱氨。这些特性与经过充分研究的APOBEC3G完全不同,APOBEC3G在长ssDNA中显示出对其靶序列的更高活性,并且靠近5'端。考虑到在与ssDNA非特异性结合后,A3B CTD的独特性质可以得到合理的解释。










































 京公网安备 11010802027423号
京公网安备 11010802027423号