当前位置:
X-MOL 学术
›
ACS Appl. Mater. Interfaces
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
DNA-Assisted Dispersion of Carbon Nanotubes and Comparison with Other Dispersing Agents
ACS Applied Materials & Interfaces ( IF 8.3 ) Pub Date : 2017-09-25 00:00:00 , DOI: 10.1021/acsami.7b06751 Debabrata Pramanik 1 , Prabal K. Maiti 1
ACS Applied Materials & Interfaces ( IF 8.3 ) Pub Date : 2017-09-25 00:00:00 , DOI: 10.1021/acsami.7b06751 Debabrata Pramanik 1 , Prabal K. Maiti 1
Affiliation
|
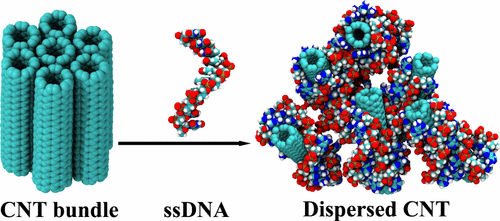
Separation and sorting of pristine carbon nanotubes (CNTs) from bundle geometry is a very challenging task due to the insoluble and nondispersive nature of CNTs in aqueous medium. Recently, many studies have been performed to address this problem using various organic and inorganic solutions, surfactant molecules, and biomolecules as dispersing agents. Recent experimental studies have reported the DNA to be highly efficient in dispersing CNTs from bundle geometry. However, there is no microscopic study and also quantitative estimation of the dispersion efficiency of the DNA. Using all-atom molecular dynamics simulation, we study the structure and stability of single-stranded DNA (ssDNA)–single-walled carbon nanotube (SWNT) (6,5) complex. To quantify the dispersion efficiency of various DNA sequences, we perform potential of mean forces (PMF) calculation between two bare SWNTs as well ssDNA-wrapped CNTs for different base sequences. From the PMF calculation, we find the PMF between two bare (6,5) SWNTs to be approximately −29 kcal/mol. For the ssDNA-wrapped SWNTs, the PMF reduces significantly and becomes repulsive. In the presence of ssDNA of different polynucleotide bases (A, T, G, and C), we present a microscopic picture of the ssDNA–SWNT (6,5) complex and also a quantitative estimate of the interaction strength between nanotubes from PMF calculation. From PMF, we show the sequence of dispersion efficiency for four different nucleic bases to be T > A > C > G. We have also presented a comparison of the dispersion efficiencies of ssDNA, flavin mononucleotide surfactant, and poly(amidoamine) (PAMAM) dendrimer by comparing their respective PMF values.
中文翻译:

碳纳米管的DNA辅助分散及其与其他分散剂的比较
由于碳纳米管在水性介质中的不溶性和非分散性,将原始碳纳米管(CNT)与束几何形状分离和分类是一项非常具有挑战性的任务。近来,已经进行了许多研究以使用各种有机和无机溶液,表面活性剂分子和生物分子作为分散剂来解决该问题。最近的实验研究报告了该DNA在分散碳纳米管束中的碳纳米管方面非常有效。然而,没有微观研究,也没有定量估计DNA的分散效率。使用全原子分子动力学模拟,我们研究了单链DNA(ssDNA)-单壁碳纳米管(SWNT)(6,5)配合物的结构和稳定性。为了量化各种DNA序列的分散效率,我们针对两个不同的碱基序列,在两个裸露的SWNT以及ssDNA包裹的CNT之间执行平均力(PMF)的计算潜力。从PMF计算中,我们发现两个裸露(6,5)SWNT之间的PMF约为-29 kcal / mol。对于包裹ssDNA的SWNT,PMF显着降低并变得排斥。在存在不同的多核苷酸碱基(A,T,G和C)的ssDNA的情况下,我们呈现了ssDNA–SWNT(6,5)复合物的显微照片,并通过PMF计算定量估算了纳米管之间的相互作用强度。从PMF中,我们显示了四种不同核酸碱基的分散效率的顺序为T> A> C>G。我们还对ssDNA,黄素单核苷酸表面活性剂,
更新日期:2017-09-26
中文翻译:

碳纳米管的DNA辅助分散及其与其他分散剂的比较
由于碳纳米管在水性介质中的不溶性和非分散性,将原始碳纳米管(CNT)与束几何形状分离和分类是一项非常具有挑战性的任务。近来,已经进行了许多研究以使用各种有机和无机溶液,表面活性剂分子和生物分子作为分散剂来解决该问题。最近的实验研究报告了该DNA在分散碳纳米管束中的碳纳米管方面非常有效。然而,没有微观研究,也没有定量估计DNA的分散效率。使用全原子分子动力学模拟,我们研究了单链DNA(ssDNA)-单壁碳纳米管(SWNT)(6,5)配合物的结构和稳定性。为了量化各种DNA序列的分散效率,我们针对两个不同的碱基序列,在两个裸露的SWNT以及ssDNA包裹的CNT之间执行平均力(PMF)的计算潜力。从PMF计算中,我们发现两个裸露(6,5)SWNT之间的PMF约为-29 kcal / mol。对于包裹ssDNA的SWNT,PMF显着降低并变得排斥。在存在不同的多核苷酸碱基(A,T,G和C)的ssDNA的情况下,我们呈现了ssDNA–SWNT(6,5)复合物的显微照片,并通过PMF计算定量估算了纳米管之间的相互作用强度。从PMF中,我们显示了四种不同核酸碱基的分散效率的顺序为T> A> C>G。我们还对ssDNA,黄素单核苷酸表面活性剂,











































 京公网安备 11010802027423号
京公网安备 11010802027423号