当前位置:
X-MOL 学术
›
RSC Med. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Integrating docking scores and key interaction profiles to improve the accuracy of molecular docking: towards novel B-RafV600E inhibitors
RSC Medicinal Chemistry ( IF 4.1 ) Pub Date : 2017-07-24 00:00:00 , DOI: 10.1039/c7md00229g Chun-Qi Hu 1, 2, 3, 4, 5 , Kang Li 1, 2, 3, 4, 5 , Ting-Ting Yao 1, 2, 3, 4, 5 , Yong-Zhou Hu 1, 2, 3, 4, 5 , Hua-Zhou Ying 1, 2, 3, 4, 5 , Xiao-Wu Dong 1, 2, 3, 4, 5
RSC Medicinal Chemistry ( IF 4.1 ) Pub Date : 2017-07-24 00:00:00 , DOI: 10.1039/c7md00229g Chun-Qi Hu 1, 2, 3, 4, 5 , Kang Li 1, 2, 3, 4, 5 , Ting-Ting Yao 1, 2, 3, 4, 5 , Yong-Zhou Hu 1, 2, 3, 4, 5 , Hua-Zhou Ying 1, 2, 3, 4, 5 , Xiao-Wu Dong 1, 2, 3, 4, 5
Affiliation
|
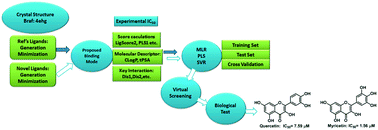
A set of ninety-eight B-RafV600E inhibitors was used for the development of a molecular docking based QSAR model using linear and non-linear regression models. The integration of docking scores and key interaction profiles significantly improved the accuracy of the QSAR models, providing reasonable statistical parameters (Rtrain2 = 0.935, Rtest2 = 0.728 and QCV2 = 0.905). The established MD-SVR (molecular docking based SMV regression) model as well as model screening of a natural product database was carried out and two natural products (quercetin and myricetin) with good prediction activities were biologically evaluated. Both compounds exhibited promising B-RafV600E inhibitory activities (ICQuercetin50 = 7.59 μM and ICMyricetin50 = 1.56 μM), suggesting a high reliability and good applicability of the established MD-SVR model in the future development of B-RafV600E inhibitors with high efficacy.
中文翻译:

整合对接分数和关键的相互作用谱以提高分子对接的准确性:针对新型B-Raf V600E抑制剂
一组九十八个B-Raf V600E抑制剂用于使用线性和非线性回归模型开发基于分子对接的QSAR模型。对接分数和关键交互配置文件的集成显着提高了QSAR模型的准确性,提供了合理的统计参数(R训练2 = 0.935,R检验2 = 0.728和Q CV 2= 0.905)。建立建立的MD-SVR(基于分子对接的SMV回归)模型以及天然产物数据库的模型筛选,并对具有良好预测活性的两种天然产物(槲皮素和杨梅素)进行了生物学评估。两种化合物均显示出有希望的B-Raf V600E抑制活性(IC槲皮素50 = 7.59μM和IC杨梅素50 = 1.56μM),表明已建立的MD-SVR模型在B-Raf V600E抑制剂的未来开发中具有很高的可靠性和良好的适用性。疗效高。
更新日期:2017-09-21
中文翻译:

整合对接分数和关键的相互作用谱以提高分子对接的准确性:针对新型B-Raf V600E抑制剂
一组九十八个B-Raf V600E抑制剂用于使用线性和非线性回归模型开发基于分子对接的QSAR模型。对接分数和关键交互配置文件的集成显着提高了QSAR模型的准确性,提供了合理的统计参数(R训练2 = 0.935,R检验2 = 0.728和Q CV 2= 0.905)。建立建立的MD-SVR(基于分子对接的SMV回归)模型以及天然产物数据库的模型筛选,并对具有良好预测活性的两种天然产物(槲皮素和杨梅素)进行了生物学评估。两种化合物均显示出有希望的B-Raf V600E抑制活性(IC槲皮素50 = 7.59μM和IC杨梅素50 = 1.56μM),表明已建立的MD-SVR模型在B-Raf V600E抑制剂的未来开发中具有很高的可靠性和良好的适用性。疗效高。











































 京公网安备 11010802027423号
京公网安备 11010802027423号