当前位置:
X-MOL 学术
›
Biochemistry
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
OleB from Bacterial Hydrocarbon Biosynthesis Is a β-Lactone Decarboxylase That Shares Key Features with Haloalkane Dehalogenases
Biochemistry ( IF 2.9 ) Pub Date : 2017-09-19 00:00:00 , DOI: 10.1021/acs.biochem.7b00667 James K. Christenson 1, 2 , Serina L. Robinson 2, 3 , Tiffany A. Engel 2 , Jack E. Richman 1, 2 , An N. Kim 2 , Larry P. Wackett 1, 2, 4
Biochemistry ( IF 2.9 ) Pub Date : 2017-09-19 00:00:00 , DOI: 10.1021/acs.biochem.7b00667 James K. Christenson 1, 2 , Serina L. Robinson 2, 3 , Tiffany A. Engel 2 , Jack E. Richman 1, 2 , An N. Kim 2 , Larry P. Wackett 1, 2, 4
Affiliation
|
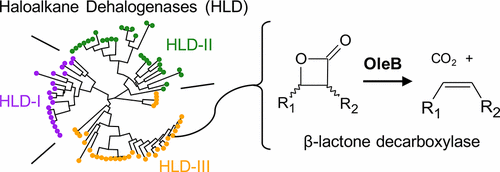
OleB is an α/β-hydrolase found in bacteria that biosynthesize long-chain olefinic hydrocarbons, but its function has remained obscure. We report that OleB from the Gram-negative bacterium Xanthomonas campestris performs an unprecedented β-lactone decarboxylation reaction, to complete cis-olefin biosynthesis. OleB reactions monitored by 1H nuclear magnetic resonance spectroscopy revealed a selectivity for decarboxylating cis-β-lactones and no discernible activity with trans-β-lactones, consistent with the known configuration of pathway intermediates. Protein sequence analyses showed OleB proteins were most related to haloalkane dehalogenases (HLDs) and retained the canonical Asp-His-Asp catalytic triad of HLDs. Unexpectedly, it was determined that an understudied subfamily, denoted as HLD-III, is comprised mostly of OleB proteins encoded within oleABCD gene clusters, suggesting a misannotation. OleB from X. campestris showed very low dehalogenase activity only against haloalkane substrates with long alkyl chains. A haloalkane substrate mimic alkylated wild-type X. campestris OleB but not OleBD114A, implicating this residue as the active site nucleophile as in HLDs. A sequence-divergent OleB, found as part of a natural OleBC fusion and classified as an HLD-III, from the Gram-positive bacterium Micrococcus luteus was demonstrated to have the same activity, stereochemical preference, and dependence on the proposed Asp nucleophile. H218O studies with M. luteus OleBC suggested that the canonical alkyl–enzyme intermediate of HLDs is hydrolyzed differently by OleB enzymes, as 18O is not incorporated into the nucleophilic aspartic acid. This work defines a previously unrecognized reaction in nature, functionally identifies some HLD-III enzymes as β-lactone decarboxylases, and posits an enzymatic mechanism of β-lactone decarboxylation.
中文翻译:

来自细菌碳氢化合物生物合成的OleB是与卤代烷脱卤酶具有关键特征的β-内酯脱羧酶
OleB是生物合成长链烯烃的细菌中发现的一种α/β水解酶,但其功能仍然不清楚。我们报道从革兰氏阴性细菌油菜黄单胞菌(Xanthomonas campestris)的OleB进行了前所未有的β-内酯脱羧反应,以完成顺式-烯烃的生物合成。通过1 H核磁共振波谱监测的OleB反应显示了对顺式-β-内酯脱羧的选择性,而反式没有可识别的活性-β-内酯,与途径中间体的已知构型一致。蛋白质序列分析显示OleB蛋白质与卤代烷脱卤酶(HLDs)最为相关,并保留了HLDs的经典Asp-His-Asp催化三联体。出乎意料的是,已确定一个未充分研究的亚家族,称为HLD-III,主要由oleABCD基因簇内编码的OleB蛋白组成,提示存在误解。来自野油菜X. Campestris的OleB仅对具有长烷基链的卤代烷底物显示出非常低的脱卤酶活性。卤代烷底物模拟烷基化的野生型喜树油菜OleB,但不模拟OleB D114A,暗示此残基是HLD中的活性位点亲核试剂。从天然革兰氏阳性细菌微球菌(Micrococcus luteus)中发现的序列差异型OleB,是天然OleBC融合的一部分,分类为HLD-III,具有相同的活性,立体化学偏好和对拟议的Asp亲核试剂的依赖性。ħ 2个18带O研究藤黄微球菌OleBC建议规范烷基-酶中间体的HLDS通过OleB酶不同水解,如18O未结合到亲核天冬氨酸中。这项工作定义了自然界以前无法识别的反应,在功能上将某些HLD-III酶鉴定为β-内酯脱羧酶,并提出了β-内酯脱羧酶机制。
更新日期:2017-09-20
中文翻译:

来自细菌碳氢化合物生物合成的OleB是与卤代烷脱卤酶具有关键特征的β-内酯脱羧酶
OleB是生物合成长链烯烃的细菌中发现的一种α/β水解酶,但其功能仍然不清楚。我们报道从革兰氏阴性细菌油菜黄单胞菌(Xanthomonas campestris)的OleB进行了前所未有的β-内酯脱羧反应,以完成顺式-烯烃的生物合成。通过1 H核磁共振波谱监测的OleB反应显示了对顺式-β-内酯脱羧的选择性,而反式没有可识别的活性-β-内酯,与途径中间体的已知构型一致。蛋白质序列分析显示OleB蛋白质与卤代烷脱卤酶(HLDs)最为相关,并保留了HLDs的经典Asp-His-Asp催化三联体。出乎意料的是,已确定一个未充分研究的亚家族,称为HLD-III,主要由oleABCD基因簇内编码的OleB蛋白组成,提示存在误解。来自野油菜X. Campestris的OleB仅对具有长烷基链的卤代烷底物显示出非常低的脱卤酶活性。卤代烷底物模拟烷基化的野生型喜树油菜OleB,但不模拟OleB D114A,暗示此残基是HLD中的活性位点亲核试剂。从天然革兰氏阳性细菌微球菌(Micrococcus luteus)中发现的序列差异型OleB,是天然OleBC融合的一部分,分类为HLD-III,具有相同的活性,立体化学偏好和对拟议的Asp亲核试剂的依赖性。ħ 2个18带O研究藤黄微球菌OleBC建议规范烷基-酶中间体的HLDS通过OleB酶不同水解,如18O未结合到亲核天冬氨酸中。这项工作定义了自然界以前无法识别的反应,在功能上将某些HLD-III酶鉴定为β-内酯脱羧酶,并提出了β-内酯脱羧酶机制。









































 京公网安备 11010802027423号
京公网安备 11010802027423号