当前位置:
X-MOL 学术
›
Anal. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
MdFDIA: A Mass Defect Based Four-Plex Data-Independent Acquisition Strategy for Proteome Quantification
Analytical Chemistry ( IF 6.7 ) Pub Date : 2017-09-19 00:00:00 , DOI: 10.1021/acs.analchem.7b01635 Yi Di , Ying Zhang , Lei Zhang , Tao Tao , Haojie Lu 1
Analytical Chemistry ( IF 6.7 ) Pub Date : 2017-09-19 00:00:00 , DOI: 10.1021/acs.analchem.7b01635 Yi Di , Ying Zhang , Lei Zhang , Tao Tao , Haojie Lu 1
Affiliation
|
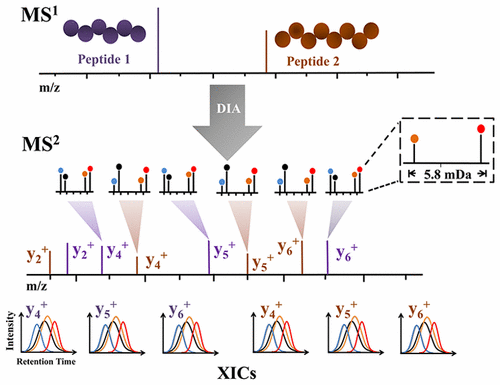
Data-independent acquisition (DIA) has recently emerged as a powerful quantitative approach for large-scale proteome quantification, providing a sensitive and reproducible alternative to data-dependent acquisition (DDA). However, lack of compatible multiplexed quantification methods is a bottleneck of DIA. To alleviate this challenge, we present a mass defect based four-plex data-independent acquisition strategy, termed “MdFDIA”, for parallel analysis of four different protein samples in a DIA experiment without the additional complexity of tandem mass spectrometry (MS2) spectra. MdFDIA is a hybrid approach that combines stable isotope labeling with amino acids in cell culture (SILAC) and dimethyl labeling. Briefly, the isotopes 13C615N2-lysine (+8.0142 Da, light) and D8-lysine (+8.0512 Da, heavy) were metabolically embedded in different proteome samples during cell culture. Then, two 13C615N2-lysine and D8-lysine labeled protein samples were digested with Lys-C, followed by in vitro labeling with light (213CD2H, +34.06312 Da) and heavy (2CD3, +34.06896 Da) dimethyl groups, respectively, producing four different pseudoisobaric labeled protein samples. The labeled samples were then equally mixed and analyzed by DIA. The subtle mass differences between the four labeled forms in MS2 scans can be resolved on an Orbitrap Fusion Lumos instrument to facilitate quantification without abundance information encoded in MS2 spectra. Additionally, a systematic investigation was carried out and revealed that MdFDIA enabled a significant decrease of the adverse impact on the accuracy of the quantitative assays arising from the chromatographic isotope effect, especially the deuterium effect, which typically occurs in a DDA experiment. Additionally, MdFDIA provided a method for validating the fragment type in the DIA spectra identification result. Furthermore, MdFDIA was applied to quantitative proteome analyses of four different breast cancer cell lines, demonstrating the feasibility of this strategy for biological applications.
中文翻译:

MdFDIA:基于质量缺陷的蛋白质组定量的四重数据独立采集策略
数据独立获取(DIA)最近成为一种强大的定量蛋白质组定量方法,为数据依赖获取(DDA)提供了一种灵敏且可重现的替代方法。但是,缺乏兼容的多元量化方法是DIA的瓶颈。为了缓解这一挑战,我们提出了一种基于质量缺陷的四重数据独立采集策略,称为“ MdFDIA”,用于在DIA实验中对四种不同蛋白质样品进行平行分析,而无需增加串联质谱(MS 2)光谱的复杂性。MdFDIA是一种混合方法,将稳定同位素标记与细胞培养物中的氨基酸(SILAC)和二甲基标记结合在一起。简而言之,同位素13 C 6 15 N在细胞培养过程中,将2-赖氨酸(+8.0142 Da,轻)和D 8-赖氨酸(+8.0512 Da,重)代谢嵌入不同的蛋白质组样品中。然后,用Lys-C消化两个13 C 6 15 N 2赖氨酸和D 8赖氨酸标记的蛋白质样品,然后分别进行轻度(2 13 CD 2 H,+34.06312 Da)和重度(2CD 3, +34.06896 Da)二甲基分别产生四个不同的伪等压标记的蛋白质样品。然后将标记的样品均等混合并通过DIA分析。MS 2中四种标记形式之间的细微质量差异扫描可以在Orbitrap Fusion Lumos仪器上进行解析,以方便定量,而无需在MS 2光谱中编码丰度信息。此外,进行了系统的研究,发现MdFDIA可以显着降低通常由DDA实验产生的色谱同位素效应(尤其是氘效应)对定量分析准确性的不利影响。另外,MdFDIA提供了一种在DIA光谱识别结果中验证片段类型的方法。此外,MdFDIA已应用于四种不同乳腺癌细胞系的定量蛋白质组分析,证明了该策略在生物学应用中的可行性。
更新日期:2017-09-20
中文翻译:

MdFDIA:基于质量缺陷的蛋白质组定量的四重数据独立采集策略
数据独立获取(DIA)最近成为一种强大的定量蛋白质组定量方法,为数据依赖获取(DDA)提供了一种灵敏且可重现的替代方法。但是,缺乏兼容的多元量化方法是DIA的瓶颈。为了缓解这一挑战,我们提出了一种基于质量缺陷的四重数据独立采集策略,称为“ MdFDIA”,用于在DIA实验中对四种不同蛋白质样品进行平行分析,而无需增加串联质谱(MS 2)光谱的复杂性。MdFDIA是一种混合方法,将稳定同位素标记与细胞培养物中的氨基酸(SILAC)和二甲基标记结合在一起。简而言之,同位素13 C 6 15 N在细胞培养过程中,将2-赖氨酸(+8.0142 Da,轻)和D 8-赖氨酸(+8.0512 Da,重)代谢嵌入不同的蛋白质组样品中。然后,用Lys-C消化两个13 C 6 15 N 2赖氨酸和D 8赖氨酸标记的蛋白质样品,然后分别进行轻度(2 13 CD 2 H,+34.06312 Da)和重度(2CD 3, +34.06896 Da)二甲基分别产生四个不同的伪等压标记的蛋白质样品。然后将标记的样品均等混合并通过DIA分析。MS 2中四种标记形式之间的细微质量差异扫描可以在Orbitrap Fusion Lumos仪器上进行解析,以方便定量,而无需在MS 2光谱中编码丰度信息。此外,进行了系统的研究,发现MdFDIA可以显着降低通常由DDA实验产生的色谱同位素效应(尤其是氘效应)对定量分析准确性的不利影响。另外,MdFDIA提供了一种在DIA光谱识别结果中验证片段类型的方法。此外,MdFDIA已应用于四种不同乳腺癌细胞系的定量蛋白质组分析,证明了该策略在生物学应用中的可行性。









































 京公网安备 11010802027423号
京公网安备 11010802027423号