当前位置:
X-MOL 学术
›
Chem. Res. Toxicol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Computational Simulations of DNA Polymerases: Detailed Insights on Structure/Function/Mechanism from Native Proteins to Cancer Variants
Chemical Research in Toxicology ( IF 3.7 ) Pub Date : 2017-09-15 00:00:00 , DOI: 10.1021/acs.chemrestox.7b00161 Alice R. Walker 1 , G. Andrés Cisneros 1
Chemical Research in Toxicology ( IF 3.7 ) Pub Date : 2017-09-15 00:00:00 , DOI: 10.1021/acs.chemrestox.7b00161 Alice R. Walker 1 , G. Andrés Cisneros 1
Affiliation
|
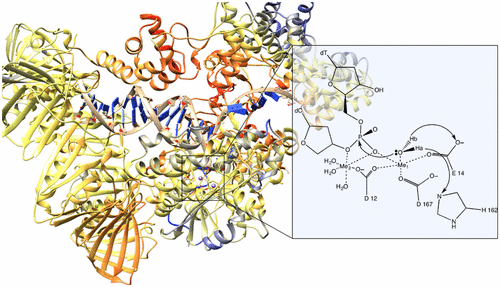
Genetic information is vital in the cell cycle of DNA-based organisms. DNA polymerases (DNA Pols) are crucial players in transactions dealing with these processes. Therefore, the detailed understanding of the structure, function, and mechanism of these proteins has been the focus of significant effort. Computational simulations have been applied to investigate various facets of DNA polymerase structure and function. These simulations have provided significant insights over the years. This perspective presents the results of various computational studies that have been employed to research different aspects of DNA polymerases including detailed reaction mechanism investigation, mutagenicity of different metal cations, possible factors for fidelity synthesis, and discovery/functional characterization of cancer-related mutations on DNA polymerases.
中文翻译:

DNA聚合酶的计算模拟:从天然蛋白质到癌症变体的结构/功能/机制的详细见解
遗传信息对于基于DNA的生物的细胞周期至关重要。DNA聚合酶(DNA Pols)在处理这些过程的交易中起着至关重要的作用。因此,对这些蛋白质的结构,功能和机制的详细了解一直是重大工作的重点。计算仿真已被用于研究DNA聚合酶结构和功能的各个方面。这些年来,这些模拟提供了重要的见识。该观点提出了已用于研究DNA聚合酶不同方面的各种计算研究的结果,包括详细的反应机理研究,不同金属阳离子的诱变性,保真合成的可能因素以及与DNA相关的癌症相关突变的发现/功能表征聚合酶。
更新日期:2017-09-15
中文翻译:

DNA聚合酶的计算模拟:从天然蛋白质到癌症变体的结构/功能/机制的详细见解
遗传信息对于基于DNA的生物的细胞周期至关重要。DNA聚合酶(DNA Pols)在处理这些过程的交易中起着至关重要的作用。因此,对这些蛋白质的结构,功能和机制的详细了解一直是重大工作的重点。计算仿真已被用于研究DNA聚合酶结构和功能的各个方面。这些年来,这些模拟提供了重要的见识。该观点提出了已用于研究DNA聚合酶不同方面的各种计算研究的结果,包括详细的反应机理研究,不同金属阳离子的诱变性,保真合成的可能因素以及与DNA相关的癌症相关突变的发现/功能表征聚合酶。











































 京公网安备 11010802027423号
京公网安备 11010802027423号