当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Toward the Standardization of Mitochondrial Proteomics: The Italian Mitochondrial Human Proteome Project Initiative
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2017-09-13 00:00:00 , DOI: 10.1021/acs.jproteome.7b00350 Tiziana Alberio 1 , Luisa Pieroni 2 , Maurizio Ronci 2, 3 , Cristina Banfi 4 , Italia Bongarzone 5 , Patrizia Bottoni 6 , Maura Brioschi 4 , Marianna Caterino 7, 8 , Clizia Chinello 9 , Antonella Cormio 10 , Flora Cozzolino 8, 11 , Vincenzo Cunsolo 12 , Simona Fontana 13 , Barbara Garavaglia 14 , Laura Giusti 15 , Viviana Greco 2 , Antonio Lucacchini 15 , Elisa Maffioli 16 , Fulvio Magni 9 , Francesca Monteleone 13 , Maria Monti 8, 11 , Valentina Monti 14 , Clara Musicco 17 , Giuseppe Petrosillo 17 , Vito Porcelli 10 , Rosaria Saletti 12 , Roberto Scatena 6 , Alessio Soggiu 16 , Gabriella Tedeschi 16, 18 , Mara Zilocchi 1 , Paola Roncada 19 , Andrea Urbani 2, 6 , Mauro Fasano 1
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2017-09-13 00:00:00 , DOI: 10.1021/acs.jproteome.7b00350 Tiziana Alberio 1 , Luisa Pieroni 2 , Maurizio Ronci 2, 3 , Cristina Banfi 4 , Italia Bongarzone 5 , Patrizia Bottoni 6 , Maura Brioschi 4 , Marianna Caterino 7, 8 , Clizia Chinello 9 , Antonella Cormio 10 , Flora Cozzolino 8, 11 , Vincenzo Cunsolo 12 , Simona Fontana 13 , Barbara Garavaglia 14 , Laura Giusti 15 , Viviana Greco 2 , Antonio Lucacchini 15 , Elisa Maffioli 16 , Fulvio Magni 9 , Francesca Monteleone 13 , Maria Monti 8, 11 , Valentina Monti 14 , Clara Musicco 17 , Giuseppe Petrosillo 17 , Vito Porcelli 10 , Rosaria Saletti 12 , Roberto Scatena 6 , Alessio Soggiu 16 , Gabriella Tedeschi 16, 18 , Mara Zilocchi 1 , Paola Roncada 19 , Andrea Urbani 2, 6 , Mauro Fasano 1
Affiliation
|
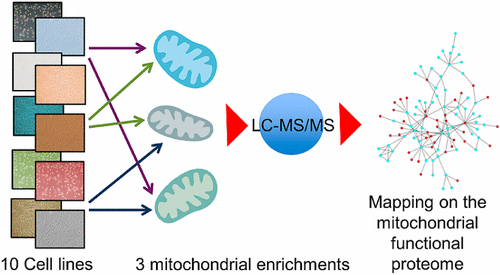
The Mitochondrial Human Proteome Project aims at understanding the function of the mitochondrial proteome and its crosstalk with the proteome of other organelles. Being able to choose a suitable and validated enrichment protocol of functional mitochondria, based on the specific needs of the downstream proteomics analysis, would greatly help the researchers in the field. Mitochondrial fractions from ten model cell lines were prepared using three enrichment protocols and analyzed on seven different LC–MS/MS platforms. All data were processed using neXtProt as reference database. The data are available for the Human Proteome Project purposes through the ProteomeXchange Consortium with the identifier PXD007053. The processed data sets were analyzed using a suite of R routines to perform a statistical analysis and to retrieve subcellular and submitochondrial localizations. Although the overall number of identified total and mitochondrial proteins was not significantly dependent on the enrichment protocol, specific line to line differences were observed. Moreover, the protein lists were mapped to a network representing the functional mitochondrial proteome, encompassing mitochondrial proteins and their first interactors. More than 80% of the identified proteins resulted in nodes of this network but with a different ability in coisolating mitochondria-associated structures for each enrichment protocol/cell line pair.
中文翻译:

致力于线粒体蛋白质组学的标准化:意大利线粒体人类蛋白质组计划计划
线粒体人类蛋白质组计划旨在了解线粒体蛋白质组的功能及其与其他细胞器蛋白质组的串扰。根据下游蛋白质组学分析的特定需求,能够选择合适且经过验证的功能性线粒体富集方案,将极大地帮助该领域的研究人员。使用三种富集方案制备了来自十个模型细胞系的线粒体级分,并在七个不同的LC-MS / MS平台上进行了分析。所有数据均使用neXtProt作为参考数据库进行处理。数据可通过ProteomeXchange联盟用于人类蛋白质组计划,其标识符为PXD007053。使用一组R例程对处理后的数据集进行分析,以进行统计分析并检索亚细胞和线粒体的定位。尽管已鉴定的总蛋白和线粒体蛋白的总数并不显着依赖于富集方案,但观察到了特定的品系差异。此外,蛋白质列表被映射到代表功能性线粒体蛋白质组的网络,其中包括线粒体蛋白质及其第一个相互作用因子。超过80%的已鉴定蛋白质产生了该网络的节点,但是对于每个富集规程/细胞系对,线粒体相关结构的共分离能力不同。尽管已鉴定的总蛋白和线粒体蛋白的总数并不显着依赖于富集方案,但观察到了特定的品系差异。此外,蛋白质列表被映射到代表功能性线粒体蛋白质组的网络,其中包括线粒体蛋白质及其第一个相互作用因子。超过80%的已鉴定蛋白质产生了该网络的节点,但是对于每个富集规程/细胞系对,线粒体相关结构的共分离能力不同。尽管已鉴定的总蛋白和线粒体蛋白的总数并不显着依赖于富集方案,但观察到了特定的品系差异。此外,蛋白质列表被映射到代表功能性线粒体蛋白质组的网络,其中包括线粒体蛋白质及其第一个相互作用因子。超过80%的已鉴定蛋白质产生了该网络的节点,但是对于每个富集规程/细胞系对,线粒体相关结构的共分离能力不同。
更新日期:2017-09-13
中文翻译:

致力于线粒体蛋白质组学的标准化:意大利线粒体人类蛋白质组计划计划
线粒体人类蛋白质组计划旨在了解线粒体蛋白质组的功能及其与其他细胞器蛋白质组的串扰。根据下游蛋白质组学分析的特定需求,能够选择合适且经过验证的功能性线粒体富集方案,将极大地帮助该领域的研究人员。使用三种富集方案制备了来自十个模型细胞系的线粒体级分,并在七个不同的LC-MS / MS平台上进行了分析。所有数据均使用neXtProt作为参考数据库进行处理。数据可通过ProteomeXchange联盟用于人类蛋白质组计划,其标识符为PXD007053。使用一组R例程对处理后的数据集进行分析,以进行统计分析并检索亚细胞和线粒体的定位。尽管已鉴定的总蛋白和线粒体蛋白的总数并不显着依赖于富集方案,但观察到了特定的品系差异。此外,蛋白质列表被映射到代表功能性线粒体蛋白质组的网络,其中包括线粒体蛋白质及其第一个相互作用因子。超过80%的已鉴定蛋白质产生了该网络的节点,但是对于每个富集规程/细胞系对,线粒体相关结构的共分离能力不同。尽管已鉴定的总蛋白和线粒体蛋白的总数并不显着依赖于富集方案,但观察到了特定的品系差异。此外,蛋白质列表被映射到代表功能性线粒体蛋白质组的网络,其中包括线粒体蛋白质及其第一个相互作用因子。超过80%的已鉴定蛋白质产生了该网络的节点,但是对于每个富集规程/细胞系对,线粒体相关结构的共分离能力不同。尽管已鉴定的总蛋白和线粒体蛋白的总数并不显着依赖于富集方案,但观察到了特定的品系差异。此外,蛋白质列表被映射到代表功能性线粒体蛋白质组的网络,其中包括线粒体蛋白质及其第一个相互作用因子。超过80%的已鉴定蛋白质产生了该网络的节点,但是对于每个富集规程/细胞系对,线粒体相关结构的共分离能力不同。











































 京公网安备 11010802027423号
京公网安备 11010802027423号