当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Scale-Invariant Biomarker Discovery in Urine and Plasma Metabolite Fingerprints
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2017-09-07 00:00:00 , DOI: 10.1021/acs.jproteome.7b00325 Helena U. Zacharias , Thorsten Rehberg , Sebastian Mehrl , Daniel Richtmann 1 , Tilo Wettig 1 , Peter J. Oefner , Rainer Spang , Wolfram Gronwald , Michael Altenbuchinger
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2017-09-07 00:00:00 , DOI: 10.1021/acs.jproteome.7b00325 Helena U. Zacharias , Thorsten Rehberg , Sebastian Mehrl , Daniel Richtmann 1 , Tilo Wettig 1 , Peter J. Oefner , Rainer Spang , Wolfram Gronwald , Michael Altenbuchinger
Affiliation
|
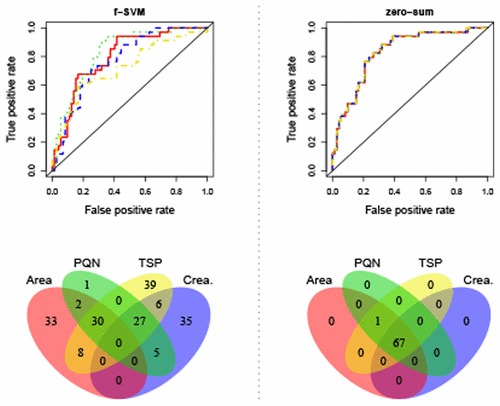
Metabolomics data is typically scaled to a common reference like a constant volume of body fluid, a constant creatinine level, or a constant area under the spectrum. Such scaling of the data, however, may affect the selection of biomarkers and the biological interpretation of results in unforeseen ways. Here, we studied how both the outcome of hypothesis tests for differential metabolite concentration and the screening for multivariate metabolite signatures are affected by the choice of scale. To overcome this problem for metabolite signatures and to establish a scale-invariant biomarker discovery algorithm, we extended linear zero-sum regression to the logistic regression framework and showed in two applications to 1H NMR-based metabolomics data how this approach overcomes the scaling problem. Logistic zero-sum regression is available as an R package as well as a high-performance computing implementation that can be downloaded at https://github.com/rehbergT/zeroSum.
中文翻译:

尿液和血浆代谢物指纹图中的尺度不变生物标志物发现
代谢组学数据通常按比例缩放到一个共同的参考标准,例如恒定的体液量,恒定的肌酐水平或光谱下的恒定面积。然而,数据的这种缩放可能以不可预见的方式影响生物标志物的选择和结果的生物学解释。在这里,我们研究了差异代谢物浓度的假设检验结果和多元代谢物特征筛选的方法如何受到规模选择的影响。为了克服代谢物签名的问题并建立尺度不变的生物标记物发现算法,我们将线性零和回归扩展到逻辑回归框架,并在两个应用中显示为1基于1 H NMR的代谢组学数据说明了该方法如何克服定标问题。Logistic零和回归可以作为R包使用,也可以从https://github.com/rehbergT/zeroSum下载高性能计算实现。
更新日期:2017-09-07
中文翻译:

尿液和血浆代谢物指纹图中的尺度不变生物标志物发现
代谢组学数据通常按比例缩放到一个共同的参考标准,例如恒定的体液量,恒定的肌酐水平或光谱下的恒定面积。然而,数据的这种缩放可能以不可预见的方式影响生物标志物的选择和结果的生物学解释。在这里,我们研究了差异代谢物浓度的假设检验结果和多元代谢物特征筛选的方法如何受到规模选择的影响。为了克服代谢物签名的问题并建立尺度不变的生物标记物发现算法,我们将线性零和回归扩展到逻辑回归框架,并在两个应用中显示为1基于1 H NMR的代谢组学数据说明了该方法如何克服定标问题。Logistic零和回归可以作为R包使用,也可以从https://github.com/rehbergT/zeroSum下载高性能计算实现。











































 京公网安备 11010802027423号
京公网安备 11010802027423号