当前位置:
X-MOL 学术
›
Biochemistry
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Nuclear Magnetic Resonance Study of RNA Structures at the 3′-End of the Hepatitis C Virus Genome
Biochemistry ( IF 2.9 ) Pub Date : 2017-09-06 00:00:00 , DOI: 10.1021/acs.biochem.7b00573 Clayton Kranawetter 1 , Samantha Brady 1 , Lizhen Sun 2 , Mark Schroeder 1 , Shi-Jie Chen 2 , Xiao Heng 1
Biochemistry ( IF 2.9 ) Pub Date : 2017-09-06 00:00:00 , DOI: 10.1021/acs.biochem.7b00573 Clayton Kranawetter 1 , Samantha Brady 1 , Lizhen Sun 2 , Mark Schroeder 1 , Shi-Jie Chen 2 , Xiao Heng 1
Affiliation
|
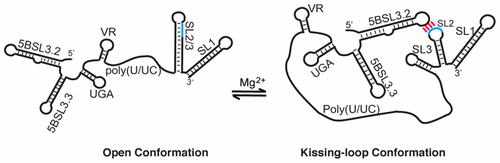
The 3′-end of the genomic RNA of the hepatitis C virus (HCV) embeds conserved elements that regulate viral RNA synthesis and protein translation by mechanisms that have yet to be elucidated. Previous studies with oligo-RNA fragments have led to multiple, mutually exclusive secondary structure predictions, indicating that HCV RNA structure may be context-dependent. Here we employed a nuclear magnetic resonance (NMR) approach that involves long-range adenosine interaction detection, coupled with site-specific 2H labeling, to probe the structure of the intact 3′-end of the HCV genome (385 nucleotides). Our data reveal that the 3′-end exists as an equilibrium mixture of two conformations: an open conformation in which the 98 nucleotides of the 3′-tail (3′X) form a two-stem–loop structure with the kissing-loop residues sequestered and a closed conformation in which the 3′X rearranges its structure and forms a long-range kissing-loop interaction with an upstream cis-acting element 5BSL3.2. The long-range kissing species is favored under high-Mg2+ conditions, and the intervening sequences do not affect the equilibrium as their secondary structures remain unchanged. The open and closed conformations are consistent with the reported function regulation of viral RNA synthesis and protein translation, respectively. Our NMR detection of these RNA conformations and the structural equilibrium in the 3′-end of the HCV genome support its roles in coordinating various steps of HCV replication.
中文翻译:

丙型肝炎病毒基因组3'-端RNA结构的核磁共振研究
丙型肝炎病毒(HCV)的基因组RNA的3'末端嵌入了保守的元件,这些元件通过尚未阐明的机制来调节病毒RNA的合成和蛋白质翻译。先前对寡RNA片段的研究导致了多个相互排斥的二级结构预测,表明HCV RNA结构可能与环境有关。在这里,我们采用了核磁共振(NMR)方法,该方法涉及远程腺苷相互作用检测,以及位点特异性2H标记,以探测HCV基因组完整的3'-末端的结构(385个核苷酸)。我们的数据表明,3'末端以两种构象的平衡混合物形式存在:一种开放构象,其中3'-尾(3'X)的98个核苷酸形成一个带有接吻环的双茎环结构残基被隔离并形成一个封闭的构象,其中3'X重新排列其结构并与上游的顺式作用元件5BSL3.2形成长距离的接环相互作用。高镁2+下,远距离接吻物种受到青睐条件,并且中间序列不影响平衡,因为它们的二级结构保持不变。打开和关闭的构象分别与报道的病毒RNA合成和蛋白质翻译的功能调节一致。我们对这些RNA构象的NMR检测和HCV基因组3'端的结构平衡证明了其在协调HCV复制各个步骤中的作用。
更新日期:2017-09-07
中文翻译:

丙型肝炎病毒基因组3'-端RNA结构的核磁共振研究
丙型肝炎病毒(HCV)的基因组RNA的3'末端嵌入了保守的元件,这些元件通过尚未阐明的机制来调节病毒RNA的合成和蛋白质翻译。先前对寡RNA片段的研究导致了多个相互排斥的二级结构预测,表明HCV RNA结构可能与环境有关。在这里,我们采用了核磁共振(NMR)方法,该方法涉及远程腺苷相互作用检测,以及位点特异性2H标记,以探测HCV基因组完整的3'-末端的结构(385个核苷酸)。我们的数据表明,3'末端以两种构象的平衡混合物形式存在:一种开放构象,其中3'-尾(3'X)的98个核苷酸形成一个带有接吻环的双茎环结构残基被隔离并形成一个封闭的构象,其中3'X重新排列其结构并与上游的顺式作用元件5BSL3.2形成长距离的接环相互作用。高镁2+下,远距离接吻物种受到青睐条件,并且中间序列不影响平衡,因为它们的二级结构保持不变。打开和关闭的构象分别与报道的病毒RNA合成和蛋白质翻译的功能调节一致。我们对这些RNA构象的NMR检测和HCV基因组3'端的结构平衡证明了其在协调HCV复制各个步骤中的作用。









































 京公网安备 11010802027423号
京公网安备 11010802027423号