PLOS Biology ( IF 7.8 ) Pub Date : 2017-09-05 , DOI: 10.1371/journal.pbio.2002458 Hakhamanesh Mostafavi , Tomaz Berisa , Felix R. Day , John R. B. Perry , Molly Przeworski , Joseph K. Pickrell
|
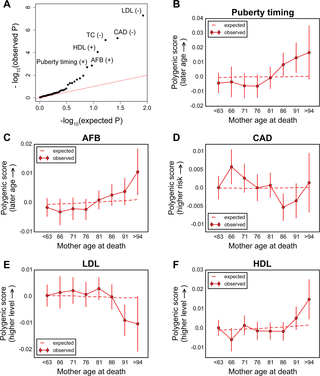
A number of open questions in human evolutionary genetics would become tractable if we were able to directly measure evolutionary fitness. As a step towards this goal, we developed a method to examine whether individual genetic variants, or sets of genetic variants, currently influence viability. The approach consists in testing whether the frequency of an allele varies across ages, accounting for variation in ancestry. We applied it to the Genetic Epidemiology Research on Adult Health and Aging (GERA) cohort and to the parents of participants in the UK Biobank. Across the genome, we found only a few common variants with large effects on age-specific mortality: tagging the APOE ε4 allele and near CHRNA3. These results suggest that when large, even late-onset effects are kept at low frequency by purifying selection. Testing viability effects of sets of genetic variants that jointly influence 1 of 42 traits, we detected a number of strong signals. In participants of the UK Biobank of British ancestry, we found that variants that delay puberty timing are associated with a longer parental life span (P~6.2 × 10−6 for fathers and P~2.0 × 10−3 for mothers), consistent with epidemiological studies. Similarly, variants associated with later age at first birth are associated with a longer maternal life span (P~1.4 × 10−3). Signals are also observed for variants influencing cholesterol levels, risk of coronary artery disease (CAD), body mass index, as well as risk of asthma. These signals exhibit consistent effects in the GERA cohort and among participants of the UK Biobank of non-British ancestry. We also found marked differences between males and females, most notably at the CHRNA3 locus, and variants associated with risk of CAD and cholesterol levels. Beyond our findings, the analysis serves as a proof of principle for how upcoming biomedical data sets can be used to learn about selection effects in contemporary humans.
中文翻译:

识别影响大型人群生存力的遗传变异
如果我们能够直接测量进化适应度,那么人类进化遗传学中的许多开放性问题将变得容易解决。为了朝这个目标迈进,我们开发了一种方法来检查单个遗传变异体或一组遗传变异体当前是否影响生存力。该方法包括测试等位基因的频率是否随年龄而变化,这要考虑到血统的变化。我们将其应用于成人健康和衰老(GERA)队列的遗传流行病学研究以及英国生物库参与者的父母。在整个基因组,我们发现年龄别死亡率大的影响只有几个常见的变异:标记的APOE ε4等位基因和近CHRNA3。这些结果表明,当大的甚至迟发的效应通过纯化选择而保持在低频率时。通过测试可共同影响42个性状之一的遗传变异集的生存力影响,我们检测到了许多强信号。在英国的祖先的英国生物银行的参与者,我们发现,该变体青春期延迟定时具有较长亲寿命相关联的(P〜6.2×10 -6为父亲和P〜2.0×10 -3对母亲),具有一致的流行病学研究。类似地,变体在第一胎都具有较长的母体寿命相关联的与后年龄(P〜1.4×10 -3)。还观察到了影响胆固醇水平,冠状动脉疾病(CAD)风险,体重指数以及哮喘风险的变异信号。这些信号在GERA队列中以及在英国非英国血统生物银行的参与者中表现出一致的效果。我们还发现男性和女性之间存在显着差异,最显着的是在CHRNA3基因座上,以及与CAD和胆固醇水平风险相关的变异。除了我们的发现外,该分析还证明了即将使用的生物医学数据集可用于了解当代人类选择效应的原理。











































 京公网安备 11010802027423号
京公网安备 11010802027423号