当前位置:
X-MOL 学术
›
Water Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A highly specific Escherichia coli qPCR and its comparison with existing methods for environmental waters
Water Research ( IF 11.4 ) Pub Date : 2017-08-25 , DOI: 10.1016/j.watres.2017.08.032 David I. Walker , Jonathan McQuillan , Michael Taiwo , Rachel Parks , Craig A. Stenton , Hywel Morgan , Matthew C. Mowlem , David N. Lees
Water Research ( IF 11.4 ) Pub Date : 2017-08-25 , DOI: 10.1016/j.watres.2017.08.032 David I. Walker , Jonathan McQuillan , Michael Taiwo , Rachel Parks , Craig A. Stenton , Hywel Morgan , Matthew C. Mowlem , David N. Lees
|
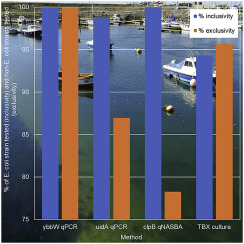
The presence of Escherichia coli in environmental waters is considered as evidence of faecal contamination and is therefore commonly used as an indicator in both water quality and food safety analysis. The long period of time between sample collection and obtaining results from existing culture based methods means that contamination events may already impact public health by the time they are detected. The adoption of molecular based methods for E. coli could significantly reduce the time to detection. A new quantitative real-time PCR (qPCR) assay was developed to detect the ybbW gene sequence, which was found to be 100% exclusive and inclusive (specific and sensitive) for E. coli and directly compared for its ability to quantify E. coli in environmental waters against colony counts, quantitative real-time NASBA (qNASBA) targeting clpB and qPCR targeting uidA. Of the 87 E. coli strains tested, 100% were found to be ybbW positive, 94.2% were culture positive, 100% were clpB positive and 98.9% were uidA positive. The qPCR assays had a linear range of quantification over several orders of magnitude, and had high amplification efficiencies when using single isolates as a template. This compared favourably with qNASBA which showed poor linearity and amplification efficiency. When the assays were applied to environmental water samples, qNASBA was unable to reliably quantify E. coli while both qPCR assays were capable of predicting E. coli concentrations in environmental waters. This study highlights the inability of qNASBA targeting mRNA to quantify E. coli in environmental waters, and presents the first E. coli qPCR assay with 100% target exclusivity. The application of a highly exclusive and inclusive qPCR assay has the potential to allow water quality managers to reliably and rapidly detect and quantify E. coli and therefore take appropriate measures to reduce the risk to public health posed by faecal contamination.
中文翻译:

高度特异性的大肠杆菌qPCR及其与现有环境水方法的比较
大肠杆菌在环境水中的存在被认为是粪便污染的证据,因此通常被用作水质和食品安全性分析的指标。从样本采集到从现有的基于文化的方法获得结果之间的时间间隔很长,这意味着污染事件在被检测到时可能已经对公共健康产生了影响。对大肠杆菌采用基于分子的方法可以大大减少检测时间。开发了一种新的定量实时PCR(qPCR)检测法来检测ybbW基因序列,该序列被发现对大肠杆菌具有100%排他性和包容性(特异性和敏感性),并直接比较了其定量能力环境水域中的大肠杆菌针对菌落计数,靶向clpB的定量实时NASBA(qNASBA)和靶向uidA的qPCR 。87个的大肠杆菌供试菌株,100%被发现是ybbW阳性,94.2%为培养阳性,100%是CLPB阳性和98.9%为的uidA阳性。qPCR测定法在多个数量级上具有线性的定量范围,并且在使用单个分离物作为模板时具有很高的扩增效率。与qNASBA相比,后者显示出较差的线性度和放大效率,因此非常有利。当将测定法应用于环境水样品时,qNASBA无法可靠地定量大肠杆菌而两种qPCR分析都能够预测环境水中的大肠杆菌浓度。这项研究突显了靶向qNASBA的mRNA无法量化环境水域中的大肠杆菌,并提出了首个具有100%目标排他性的大肠杆菌qPCR检测方法。高度排他性和包容性qPCR分析的应用可能使水质管理者能够可靠,快速地检测和定量大肠杆菌,因此采取适当措施降低粪便污染对公共健康的风险。
更新日期:2017-08-25
中文翻译:

高度特异性的大肠杆菌qPCR及其与现有环境水方法的比较
大肠杆菌在环境水中的存在被认为是粪便污染的证据,因此通常被用作水质和食品安全性分析的指标。从样本采集到从现有的基于文化的方法获得结果之间的时间间隔很长,这意味着污染事件在被检测到时可能已经对公共健康产生了影响。对大肠杆菌采用基于分子的方法可以大大减少检测时间。开发了一种新的定量实时PCR(qPCR)检测法来检测ybbW基因序列,该序列被发现对大肠杆菌具有100%排他性和包容性(特异性和敏感性),并直接比较了其定量能力环境水域中的大肠杆菌针对菌落计数,靶向clpB的定量实时NASBA(qNASBA)和靶向uidA的qPCR 。87个的大肠杆菌供试菌株,100%被发现是ybbW阳性,94.2%为培养阳性,100%是CLPB阳性和98.9%为的uidA阳性。qPCR测定法在多个数量级上具有线性的定量范围,并且在使用单个分离物作为模板时具有很高的扩增效率。与qNASBA相比,后者显示出较差的线性度和放大效率,因此非常有利。当将测定法应用于环境水样品时,qNASBA无法可靠地定量大肠杆菌而两种qPCR分析都能够预测环境水中的大肠杆菌浓度。这项研究突显了靶向qNASBA的mRNA无法量化环境水域中的大肠杆菌,并提出了首个具有100%目标排他性的大肠杆菌qPCR检测方法。高度排他性和包容性qPCR分析的应用可能使水质管理者能够可靠,快速地检测和定量大肠杆菌,因此采取适当措施降低粪便污染对公共健康的风险。











































 京公网安备 11010802027423号
京公网安备 11010802027423号