Cell Host & Microbe ( IF 20.6 ) Pub Date : 2017-08-17 , DOI: 10.1016/j.chom.2017.07.016 Nora C. Pyenson , Kaitlyn Gayvert , Andrew Varble , Olivier Elemento , Luciano A. Marraffini
|
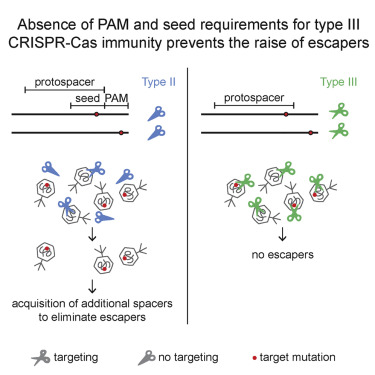
CRISPR loci are a cluster of repeats separated by short “spacer” sequences derived from prokaryotic viruses and plasmids that determine the targets of the host’s CRISPR-Cas immune response against its invaders. For type I and II CRISPR-Cas systems, single-nucleotide mutations in the seed or protospacer adjacent motif (PAM) of the target sequence cause immune failure and allow viral escape. This is overcome by the acquisition of multiple spacers that target the same invader. Here we show that targeting by the Staphylococcus epidermidis type III-A CRISPR-Cas system does not require PAM or seed sequences, and thus prevents viral escape via single-nucleotide substitutions. Instead, viral escapers can only arise through complete target deletion. Our work shows that, as opposed to type I and II systems, the relaxed specificity of type III CRISPR-Cas targeting provides robust immune responses that can lead to viral extinction with a single spacer targeting an essential phage sequence.
中文翻译:

细菌III型CRISPR-Cas免疫过程中的广泛靶向特异性可限制病毒逃逸
CRISPR基因座是由原核病毒和质粒衍生的短“间隔”序列分隔的重复序列簇,这些序列决定了宿主针对其入侵者的CRISPR-Cas免疫反应的靶标。对于I型和II型CRISPR-Cas系统,靶序列的种子或原间隔子相邻基序(PAM)中的单核苷酸突变会导致免疫衰竭并允许病毒逃逸。通过获取针对同一入侵者的多个间隔物可以克服这一问题。在这里,我们显示了由表皮葡萄球菌靶向III-A型CRISPR-Cas系统不需要PAM或种子序列,因此可以防止通过单核苷酸取代引起的病毒逃逸。取而代之的是,病毒逃逸者只能通过完整的靶标缺失来产生。我们的工作表明,与I型和II型系统相反,III型CRISPR-Cas靶向的宽松特异性提供了强大的免疫反应,可通过靶向基本噬菌体序列的单个间隔子导致病毒灭绝。









































 京公网安备 11010802027423号
京公网安备 11010802027423号