当前位置:
X-MOL 学术
›
ACS Chem. Neurosci.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Molecular Dynamics Simulation Study on the Binding and Stabilization Mechanism of Antiprion Compounds to the “Hot Spot” Region of PrPC
ACS Chemical Neuroscience ( IF 4.1 ) Pub Date : 2017-08-22 00:00:00 , DOI: 10.1021/acschemneuro.7b00214 Shuangyan Zhou 1 , Xuewei Liu 2 , Xiaoli An 2 , Xiaojun Yao 2, 3 , Huanxiang Liu 1
ACS Chemical Neuroscience ( IF 4.1 ) Pub Date : 2017-08-22 00:00:00 , DOI: 10.1021/acschemneuro.7b00214 Shuangyan Zhou 1 , Xuewei Liu 2 , Xiaoli An 2 , Xiaojun Yao 2, 3 , Huanxiang Liu 1
Affiliation
|
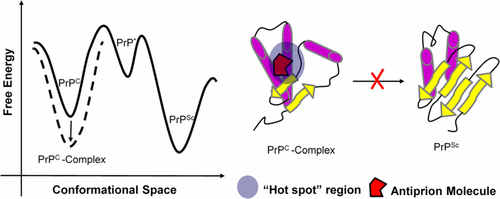
Structural transitions in the prion protein from the cellular form, PrPC, into the pathological isoform, PrPSc, are regarded as the main cause of the transmissible spongiform encephalopathies, also known as prion diseases. Hence, discovering and designing effective antiprion drugs that can inhibit PrPC to PrPSc conversion is regarded as a promising way to cure prion disease. Among several strategies to inhibit PrPC to PrPSc conversion, stabilizing the native PrPC via specific binding is believed to be one of the valuable approaches and many antiprion compounds have been reported based on this strategy. However, the detailed mechanism to stabilize the native PrPC is still unknown. As such, to unravel the stabilizing mechanism of these compounds to PrPC is valuable for the further design and discovery of antiprion compounds. In this study, by molecular dynamics simulation method, we investigated the stabilizing mechanism of several antiprion compounds on PrPC that were previously reported to have specific binding to the “hot spot” region of PrPC. Our simulation results reveal that the stabilization mechanism of specific binding compounds can be summarized as (I) to stabilize both the flexible C-terminal of α2 and the hydrophobic core, such as BMD42-29 and GN8; (II) to stabilize the hydrophobic core, such as J1 and GJP49; (III) to stabilize the overall structure of PrPC by high binding affinity, as NPR-056. In addition, as indicated by the H-bond analysis and decomposition analysis of binding free energy, the residues N159 and Q160 play an important role in the specific binding of the studied compounds and all these compounds interact with PrPC in a similar way with the key interacting residues L130 in the β1 strand, P158, N159, Q160, etc. in the α1-β2 loop, and H187, T190, T191, etc. in the α2 C-terminus although the compounds have large structural difference. As a whole, our obtained results can provide some insights into the specific binding mechanism of main antiprion compounds to the “hot spot” region of PrPC at the molecular level and also provide guidance for effective antiprion drug design in the future.
中文翻译:

Antiprion化合物对PrP C “热点”区域的结合和稳定机理的分子动力学模拟研究
ion病毒蛋白从细胞形式PrP C到病理亚型PrP Sc的结构转变被认为是可传播的海绵状脑病的主要诱因,也被称为ion病毒疾病。因此,发现和设计可以抑制PrP C到PrP Sc转化的有效抗pr病毒药物被认为是治愈病毒疾病的一种有前途的方法。在几种抑制PrP C到PrP Sc转化,稳定天然PrP C的策略中通过特异性结合的方法被认为是有价值的方法之一,并且已经基于该策略报道了许多抗pr病毒化合物。然而,稳定天然PrP C的详细机制仍是未知的。这样,揭示这些化合物对PrP C的稳定机理对于进一步设计和发现抗compounds病毒化合物是有价值的。在这项研究中,通过分子动力学模拟方法,我们研究了对朊病毒几个抗朊病毒化合物的稳定机构Ç先前报道具有特异性结合的PrP的“热点”区域Ç。我们的模拟结果表明,特异性结合化合物的稳定机制可以概括为:(I)稳定α2的柔性C端和疏水性核(例如BMD42-29和GN8);(II)稳定疏水核,例如J1和GJP49;(III)以稳定的PrP的整体结构Ç由高结合亲和力,如NPR-056。此外,如结合自由能的H键分析和分解分析所示,残基N159和Q160在所研究化合物的特异性结合中起着重要作用,并且所有这些化合物均与PrP C相互作用。以相同的方式与β1链中的关键相互作用残基L130,α1-β2环中的P158,N159,Q160等以及α2C端中的H187,T190,T191等相似,尽管这些化合物具有较大的结构差异。总体而言,我们获得的结果可为主要抗pr病毒化合物在分子水平上与PrP C “热点”区域的特异性结合机理提供一些见识,并为将来有效的抗pr病毒药物设计提供指导。
更新日期:2017-08-23
中文翻译:

Antiprion化合物对PrP C “热点”区域的结合和稳定机理的分子动力学模拟研究
ion病毒蛋白从细胞形式PrP C到病理亚型PrP Sc的结构转变被认为是可传播的海绵状脑病的主要诱因,也被称为ion病毒疾病。因此,发现和设计可以抑制PrP C到PrP Sc转化的有效抗pr病毒药物被认为是治愈病毒疾病的一种有前途的方法。在几种抑制PrP C到PrP Sc转化,稳定天然PrP C的策略中通过特异性结合的方法被认为是有价值的方法之一,并且已经基于该策略报道了许多抗pr病毒化合物。然而,稳定天然PrP C的详细机制仍是未知的。这样,揭示这些化合物对PrP C的稳定机理对于进一步设计和发现抗compounds病毒化合物是有价值的。在这项研究中,通过分子动力学模拟方法,我们研究了对朊病毒几个抗朊病毒化合物的稳定机构Ç先前报道具有特异性结合的PrP的“热点”区域Ç。我们的模拟结果表明,特异性结合化合物的稳定机制可以概括为:(I)稳定α2的柔性C端和疏水性核(例如BMD42-29和GN8);(II)稳定疏水核,例如J1和GJP49;(III)以稳定的PrP的整体结构Ç由高结合亲和力,如NPR-056。此外,如结合自由能的H键分析和分解分析所示,残基N159和Q160在所研究化合物的特异性结合中起着重要作用,并且所有这些化合物均与PrP C相互作用。以相同的方式与β1链中的关键相互作用残基L130,α1-β2环中的P158,N159,Q160等以及α2C端中的H187,T190,T191等相似,尽管这些化合物具有较大的结构差异。总体而言,我们获得的结果可为主要抗pr病毒化合物在分子水平上与PrP C “热点”区域的特异性结合机理提供一些见识,并为将来有效的抗pr病毒药物设计提供指导。











































 京公网安备 11010802027423号
京公网安备 11010802027423号