当前位置:
X-MOL 学术
›
ChemBioChem
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Analysis of Structure–Activity Relationships Based on the Hepatitis C Virus SLIIb Internal Ribosomal Entry Sequence RNA-Targeting GGHYRFK⋅Cu Complex
ChemBioChem ( IF 2.6 ) Pub Date : 2017-08-07 06:41:43 , DOI: 10.1002/cbic.201700228 Martin James Ross 1, 2 , Insiya Fidai 1, 3 , James A. Cowan 1, 3, 4, 5
ChemBioChem ( IF 2.6 ) Pub Date : 2017-08-07 06:41:43 , DOI: 10.1002/cbic.201700228 Martin James Ross 1, 2 , Insiya Fidai 1, 3 , James A. Cowan 1, 3, 4, 5
Affiliation
|
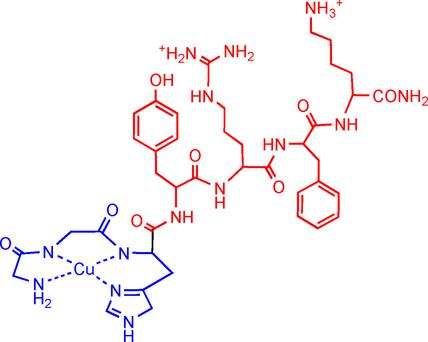
New therapeutics for targeting the hepatitis C virus (HCV) have been released in recent years. Although they are less prone to resistance, they are still administered in cocktails as a combination of drugs targeting various aspects of the viral life cycle. Herein, we aim to contribute to an arsenal of new HCV therapeutics by targeting the HCV internal ribosomal entry sequence (IRES) RNA through the development of catalytic metallodrugs that function to degrade rather than inhibit the target molecule. Based on a previously characterized HCV IRES stem-loop IIb RNA-targeting metallopeptide Cu-GGHYrFK (1⋅Cu), an all-l analogue (3⋅Cu) and a series of additional complexes with single alanine substitutions in the targeting domain were prepared and screened to determine the influence each amino acid side chain on RNA localization and recognition, and catalytic reactivity toward the RNA. Additional substitutions of the tyrosine position in complex 3⋅Cu were also investigated. Good agreement between calculated and measured binding affinities provided support for in silico modeling of the SLIIb RNA binding site and correlations with RNA cleavage sites. Examination of the cleavage products from reaction of the Cu complexes with SLIIb provided mechanistic insights, with the first observation of the 5′-geminal diol and 5′-phosphopropenal as products through the use of a Cu⋅ATCUN catalytic motif. Together, the data yielded insights into structure–function relationships that will guide future optimization efforts.
中文翻译:

基于丙型肝炎病毒SLIIb内部核糖体进入序列RNA靶向GGHYRFK·Cu复合物的结构与活性关系分析
近年来,已经发布了针对丙型肝炎病毒(HCV)的新疗法。尽管它们不易产生耐药性,但它们仍以鸡尾酒形式作为针对病毒生命周期各个方面的药物组合给药。在本文中,我们旨在通过开发催化金属药物来发挥作用,以开发HCV内部核糖体进入序列(IRES)RNA,以催化其降解而不是抑制靶标分子,从而为HCV新疗法做出贡献。基于先前鉴定的HCV IRES茎环IIb RNA靶向金属肽Cu-GGHYrFK(1⋅Cu),是全l类似物(3⋅制备并筛选在靶向结构域中具有单个丙氨酸取代的一系列Cu(1)和其他附加的复合物,以确定每个氨基酸侧链对RNA定位和识别的影响以及对RNA的催化反应性。还研究了复合物3⋅Cu中酪氨酸位置的其他取代。计算和测量的结合亲和力之间的良好一致性为SLIIb RNA结合位点的计算机模拟以及与RNA裂解位点的相关性提供了支持。从铜配合物SLIIb的反应裂解产物的检查提供机械的见解,通过使用Cu的5'-偕二醇和5'-phosphopropenal作为产品的第一次观察⋅ATCUN催化基序。在一起,这些数据产生了对结构与功能关系的深刻见解,将指导未来的优化工作。
更新日期:2017-08-07
中文翻译:

基于丙型肝炎病毒SLIIb内部核糖体进入序列RNA靶向GGHYRFK·Cu复合物的结构与活性关系分析
近年来,已经发布了针对丙型肝炎病毒(HCV)的新疗法。尽管它们不易产生耐药性,但它们仍以鸡尾酒形式作为针对病毒生命周期各个方面的药物组合给药。在本文中,我们旨在通过开发催化金属药物来发挥作用,以开发HCV内部核糖体进入序列(IRES)RNA,以催化其降解而不是抑制靶标分子,从而为HCV新疗法做出贡献。基于先前鉴定的HCV IRES茎环IIb RNA靶向金属肽Cu-GGHYrFK(1⋅Cu),是全l类似物(3⋅制备并筛选在靶向结构域中具有单个丙氨酸取代的一系列Cu(1)和其他附加的复合物,以确定每个氨基酸侧链对RNA定位和识别的影响以及对RNA的催化反应性。还研究了复合物3⋅Cu中酪氨酸位置的其他取代。计算和测量的结合亲和力之间的良好一致性为SLIIb RNA结合位点的计算机模拟以及与RNA裂解位点的相关性提供了支持。从铜配合物SLIIb的反应裂解产物的检查提供机械的见解,通过使用Cu的5'-偕二醇和5'-phosphopropenal作为产品的第一次观察⋅ATCUN催化基序。在一起,这些数据产生了对结构与功能关系的深刻见解,将指导未来的优化工作。









































 京公网安备 11010802027423号
京公网安备 11010802027423号