当前位置:
X-MOL 学术
›
Integr. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A computational framework for distinguishing direct versus indirect interactions in human functional protein-protein interaction networks.
Integrative Biology ( IF 1.5 ) Pub Date : 2017-07-17 , DOI: 10.1039/c7ib00013h Suyu Mei 1 , Erik K Flemington , Kun Zhang
Integrative Biology ( IF 1.5 ) Pub Date : 2017-07-17 , DOI: 10.1039/c7ib00013h Suyu Mei 1 , Erik K Flemington , Kun Zhang
Affiliation
|
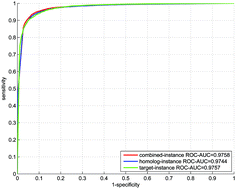
Recognition of indirect interactions is instrumental to in silico reconstruction of signaling pathways and sheds light on the exploration of unknown physical paths between two indirectly interacting genes. However, very limited computational methods have explicitly exploited the indirect interactions with experimental evidence thus far. In this work, we attempt to distinguish direct versus indirect interactions in human functional protein-protein interaction (PPI) networks via a predictive l2-regularized logistic regression model built on the experimental data. The l2-regularized logistic regression method is adopted to counteract the potential homolog noise and reduce the computational complexity on large training data. Computational results show that the proposed model demonstrates promising performance even though the training data are highly skewed. From the 304 799 PPIs that are curated in several databases, the proposed method detects 23 131 indirect interactions, most of which have been verified by the breadth-first graph search algorithm to find dozens of physical paths between the interacting partners. Pathway enrichment analysis shows that most of the physical paths can be mapped onto more than one human signaling pathway, indicating that there do exist a series of biochemical signals between the two indirectly interacting genes. The interactome-scale computational results promise to provide useful cues to the following applications: (1) exploration of unknown physical PPIs or physical paths between two indirectly interacting genes; (2) amending or extending the existing signaling pathways; (3) recognition of the physical PPIs for druggable target discovery.
中文翻译:

一个计算框架,用于区分人类功能性蛋白质-蛋白质相互作用网络中的直接相互作用和间接相互作用。
间接相互作用的识别有助于计算机重建信号通路,并为探索两个间接相互作用的基因之间未知的物理途径提供了启示。但是,到目前为止,非常有限的计算方法已明确利用了与实验证据之间的间接相互作用。在这项工作中,我们尝试通过建立在实验数据基础上的预测性L2正规化Logistic回归模型来区分人类功能性蛋白质-蛋白质相互作用(PPI)网络中的直接相互作用和间接相互作用。采用l2正则化logistic回归方法来抵消潜在的同源噪声并降低大训练数据上的计算复杂性。计算结果表明,即使训练数据高度偏斜,所提出的模型也显示出令人鼓舞的性能。从在多个数据库中管理的304799个PPI中,所提出的方法检测到23131个间接交互,其中大多数已通过广度优先图搜索算法进行了验证,以找到交互伙伴之间的数十条物理路径。途径富集分析表明,大多数物理途径可以映射到多个人信号传导途径上,这表明在两个间接相互作用的基因之间确实存在一系列生化信号。相互作用组规模的计算结果有望为以下应用提供有用的线索:(1)探索未知的物理PPI或两个间接相互作用的基因之间的物理路径;(2)修改或扩展现有的信号通路;(3)识别用于药物目标的物理PPI。
更新日期:2017-05-19
中文翻译:

一个计算框架,用于区分人类功能性蛋白质-蛋白质相互作用网络中的直接相互作用和间接相互作用。
间接相互作用的识别有助于计算机重建信号通路,并为探索两个间接相互作用的基因之间未知的物理途径提供了启示。但是,到目前为止,非常有限的计算方法已明确利用了与实验证据之间的间接相互作用。在这项工作中,我们尝试通过建立在实验数据基础上的预测性L2正规化Logistic回归模型来区分人类功能性蛋白质-蛋白质相互作用(PPI)网络中的直接相互作用和间接相互作用。采用l2正则化logistic回归方法来抵消潜在的同源噪声并降低大训练数据上的计算复杂性。计算结果表明,即使训练数据高度偏斜,所提出的模型也显示出令人鼓舞的性能。从在多个数据库中管理的304799个PPI中,所提出的方法检测到23131个间接交互,其中大多数已通过广度优先图搜索算法进行了验证,以找到交互伙伴之间的数十条物理路径。途径富集分析表明,大多数物理途径可以映射到多个人信号传导途径上,这表明在两个间接相互作用的基因之间确实存在一系列生化信号。相互作用组规模的计算结果有望为以下应用提供有用的线索:(1)探索未知的物理PPI或两个间接相互作用的基因之间的物理路径;(2)修改或扩展现有的信号通路;(3)识别用于药物目标的物理PPI。











































 京公网安备 11010802027423号
京公网安备 11010802027423号