当前位置:
X-MOL 学术
›
J. Am. Chem. Soc.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
An Assay Based on SAMDI Mass Spectrometry for Profiling Protein Interaction Domains
Journal of the American Chemical Society ( IF 14.4 ) Pub Date : 2017-07-21 00:00:00 , DOI: 10.1021/jacs.7b03805 Patrick T. O’Kane 1 , Milan Mrksich 1
Journal of the American Chemical Society ( IF 14.4 ) Pub Date : 2017-07-21 00:00:00 , DOI: 10.1021/jacs.7b03805 Patrick T. O’Kane 1 , Milan Mrksich 1
Affiliation
|
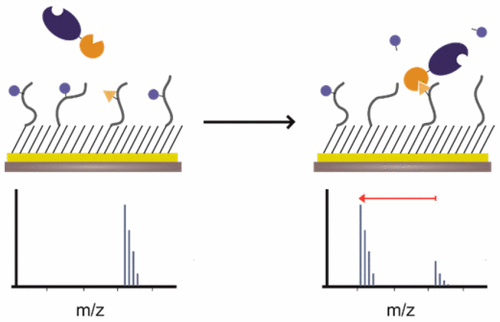
This paper describes an assay that can profile the binding of a protein to ligands and can rank the affinities of a library of ligands. The method is based on the enhanced rate of an enzyme-mediated reaction that follows from colocalization of the enzyme and substrate by a protein–ligand interaction. This assay uses a self-assembled monolayer that presents a candidate peptide ligand for a receptor and a peptide substrate for an enzyme. The receptor is prepared as a fusion to the relevant enzyme so that binding of the receptor to the immobilized ligand brings the enzyme to the surface, where it can more rapidly modify its substrate. The extent of conversion of the substrate to product is therefore a measure of the average time the ligand–receptor complex is present and is quantified using the SAMDI mass spectrometry technique. The approach is used to profile the binding of chromodomain proteins to methylated lysine peptides derived from the histone 3 protein. The relative affinities for the peptide ligands found in this work agreed with results from prior studies. Additionally, this work revealed cross-talk interactions whereby phosphorylation of certain residues impaired binding of chromodomains to the peptide ligands. The method presented here, which we term protein interaction by SAMDI (PI-SAMDI), has the advantages that it is applicable to low-affinity interactions because the complexes are not observed directly, but rather leave a “covalent record” of the interaction that is measured with mass spectrometry and because it is compatible with laboratory automation for high-throughput analysis.
中文翻译:

基于SAMDI质谱的蛋白质相互作用域分析
本文介绍了一种可以分析蛋白质与配体的结合并可以对配体文库的亲和力进行分类的测定方法。该方法基于通过蛋白质-配体相互作用使酶和底物共定位,从而提高了酶介导的反应速率。该测定使用自组装的单层,该单层提供了受体的候选肽配体和酶的肽底物。受体被制备为与相关酶的融合体,因此受体与固定配体的结合将酶带到表面,在此表面可以更快速地修饰其底物。因此,底物向产物的转化程度是配体-受体复合物存在的平均时间的量度,并使用SAMDI质谱技术进行定量。该方法用于分析色域蛋白与衍生自组蛋白3蛋白的甲基化赖氨酸肽的结合。在这项工作中发现的肽配体的相对亲和力与以前的研究结果相吻合。另外,这项工作揭示了串扰相互作用,由此某些残基的磷酸化削弱了染色体结构域与肽配体的结合。这里介绍的方法(我们称其为通过SAMDI进行蛋白质相互作用(PI-SAMDI))的优点是,它适用于低亲和力相互作用,因为不会直接观察到复合物,而是留下了相互作用的“共价记录”质谱法可通过质谱法进行测量,因为它与实验室自动化兼容,可进行高通量分析。
更新日期:2017-07-22
中文翻译:

基于SAMDI质谱的蛋白质相互作用域分析
本文介绍了一种可以分析蛋白质与配体的结合并可以对配体文库的亲和力进行分类的测定方法。该方法基于通过蛋白质-配体相互作用使酶和底物共定位,从而提高了酶介导的反应速率。该测定使用自组装的单层,该单层提供了受体的候选肽配体和酶的肽底物。受体被制备为与相关酶的融合体,因此受体与固定配体的结合将酶带到表面,在此表面可以更快速地修饰其底物。因此,底物向产物的转化程度是配体-受体复合物存在的平均时间的量度,并使用SAMDI质谱技术进行定量。该方法用于分析色域蛋白与衍生自组蛋白3蛋白的甲基化赖氨酸肽的结合。在这项工作中发现的肽配体的相对亲和力与以前的研究结果相吻合。另外,这项工作揭示了串扰相互作用,由此某些残基的磷酸化削弱了染色体结构域与肽配体的结合。这里介绍的方法(我们称其为通过SAMDI进行蛋白质相互作用(PI-SAMDI))的优点是,它适用于低亲和力相互作用,因为不会直接观察到复合物,而是留下了相互作用的“共价记录”质谱法可通过质谱法进行测量,因为它与实验室自动化兼容,可进行高通量分析。











































 京公网安备 11010802027423号
京公网安备 11010802027423号