当前位置:
X-MOL 学术
›
Acc. Chem. Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Sensing Mechanisms in the Redox-Regulated, [2Fe–2S] Cluster-Containing, Bacterial Transcriptional Factor SoxR
Accounts of Chemical Research ( IF 16.4 ) Pub Date : 2017-06-21 00:00:00 , DOI: 10.1021/acs.accounts.7b00137 Kazuo Kobayashi 1
Accounts of Chemical Research ( IF 16.4 ) Pub Date : 2017-06-21 00:00:00 , DOI: 10.1021/acs.accounts.7b00137 Kazuo Kobayashi 1
Affiliation
|
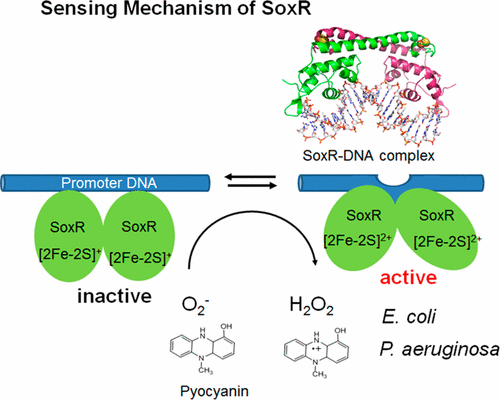
Bacteria possess molecular biosensors that enable responses to a variety of stressful conditions, including oxidative stress, toxic compounds, and interactions with other organisms, through elaborately coordinated regulation of gene expression. In Escherichia coli and related bacteria, the transcription factor SoxR functions as a sensor of oxidative stress and nitric oxide (NO). SoxR protein contains a [2Fe–2S] cluster essential for its transcription-enhancing activity, which is regulated by redox changes in the [2Fe–2S] cluster. We have explored the mechanistic and structural basis of SoxR proteins function and determined how the chemistry at the [2Fe–2S] cluster causes the subsequent regulatory response. In this Account, I describe our recent achievements in three different areas using physicochemical techniques, primarily pulse radiolysis. First, redox-dependent conformational changes in SoxR-bound DNA were studied by site-specifically replacing selected bases with the fluorescent probes 2-aminopurine and pyrrolocytosine. X-ray analyses of the DNA-SoxR complex in the oxidized state revealed that the DNA structure is distorted in the center regions, resulting in local untwisting of base pairs. However, the inactive, reduced state had remained uncharacterized. We found that reduction of the [2Fe–2S] cluster in the SoxR-DNA complex weakens the fluorescence intensity within a region confined to the central base pairs in the promoter region. Second, the reactions of NO with [2Fe–2S] clusters of E. coli SoxR were analyzed using pulse radiolysis. The transcriptional activation of SoxR in E. coli occurs through direct modification of [2Fe–2S] by NO to form a dinitrosyl iron complex (DNIC). The reaction of NO with [2Fe–2S] cluster of SoxR proceeded nearly quantitatively with concomitant reductive elimination of two equivalents S0 atoms. Intermediate nitrosylation products, however, were too unstable to observe. We found that the conversion proceeds through at least two steps, with the faster phase being the first reaction of the NO molecule with the [2Fe–2S] cluster. The slower reaction with the second equivalent NO molecule, however, was important for the formation of DNIC. Third, to elucidate the differences between the distinct responses of SoxR proteins from two different species, we studied the interaction of E. coli and Pseudomonas aeruginosa SoxR with superoxide anion using a mutagenic approach. Despite the homology between E. coli SoxR and P. aeruginosa SoxR, the function of P. aeruginosa SoxR differs from that of E. coli. The substitution of E. coli SoxR lysine residues, located close to [2Fe–2S] clusters, into P. aeruginosa SoxR dramatically affected the reaction with superoxide anion.
中文翻译:

氧化还原调节的,[2Fe–2S]群集,细菌转录因子SoxR中的传感机制。
细菌拥有分子生物传感器,可以通过精心协调地调节基因表达,从而对多种应激条件做出反应,包括氧化应激,有毒化合物以及与其他生物的相互作用。在大肠杆菌中以及相关细菌,转录因子SoxR可以作为氧化应激和一氧化氮(NO)的传感器。SoxR蛋白包含一个[2Fe–2S]簇,对于其转录增强活性至关重要,该簇受[2Fe–2S]簇中的氧化还原变化的调节。我们已经探索了SoxR蛋白质功能的机制和结构基础,并确定了[2Fe–2S]簇上的化学反应如何引起随后的调节反应。在本报告中,我将介绍使用物理化学技术(主要是脉冲辐射分解)在三个不同领域的最新成就。首先,通过用荧光探针2-氨基嘌呤和吡咯并胞嘧啶位点特异性取代选定的碱基,研究了SoxR结合的DNA中氧化还原依赖性的构象变化。对处于氧化状态的DNA-SoxR复合物的X射线分析表明,DNA结构在中心区域变形,导致碱基对局部解捻。但是,未激活的还原状态仍未表征。我们发现,SoxR-DNA复合物中[2Fe–2S]簇的减少减弱了局限于启动子区域中央碱基对的区域内的荧光强度。其次,NO与[2Fe–2S]团簇的反应大肠杆菌SoxR使用脉冲放射分析法进行了分析。SoxR在大肠杆菌中的转录激活是通过NO对[2Fe-2S]的直接修饰而形成的,形成二亚硝基铁络合物(DNIC)。NO与SoxR的[2Fe–2S]簇的反应几乎定量进行,同时还原消除了两个当量S 0原子。但是,中间的亚硝基化产物太不稳定而无法观察到。我们发现转化过程至少要经过两个步骤,其中较快的阶段是NO分子与[2Fe–2S]团簇的第一个反应。但是,与第二当量NO分子的反应较慢,对于DNIC的形成很重要。第三,为了阐明两种不同物种的SoxR蛋白不同反应之间的差异,我们使用诱变方法研究了大肠杆菌和铜绿假单胞菌SoxR与超氧阴离子的相互作用。尽管之间的同源性的大肠杆菌SoxR和铜绿假单胞菌SoxR,功能绿脓杆菌SoxR不同于的大肠杆菌。的取代大肠杆菌SoxR赖氨酸残基,位置靠近的[2Fe-2S]簇,到铜绿假单胞菌SoxR显着影响与超氧阴离子反应。
更新日期:2017-06-21
中文翻译:

氧化还原调节的,[2Fe–2S]群集,细菌转录因子SoxR中的传感机制。
细菌拥有分子生物传感器,可以通过精心协调地调节基因表达,从而对多种应激条件做出反应,包括氧化应激,有毒化合物以及与其他生物的相互作用。在大肠杆菌中以及相关细菌,转录因子SoxR可以作为氧化应激和一氧化氮(NO)的传感器。SoxR蛋白包含一个[2Fe–2S]簇,对于其转录增强活性至关重要,该簇受[2Fe–2S]簇中的氧化还原变化的调节。我们已经探索了SoxR蛋白质功能的机制和结构基础,并确定了[2Fe–2S]簇上的化学反应如何引起随后的调节反应。在本报告中,我将介绍使用物理化学技术(主要是脉冲辐射分解)在三个不同领域的最新成就。首先,通过用荧光探针2-氨基嘌呤和吡咯并胞嘧啶位点特异性取代选定的碱基,研究了SoxR结合的DNA中氧化还原依赖性的构象变化。对处于氧化状态的DNA-SoxR复合物的X射线分析表明,DNA结构在中心区域变形,导致碱基对局部解捻。但是,未激活的还原状态仍未表征。我们发现,SoxR-DNA复合物中[2Fe–2S]簇的减少减弱了局限于启动子区域中央碱基对的区域内的荧光强度。其次,NO与[2Fe–2S]团簇的反应大肠杆菌SoxR使用脉冲放射分析法进行了分析。SoxR在大肠杆菌中的转录激活是通过NO对[2Fe-2S]的直接修饰而形成的,形成二亚硝基铁络合物(DNIC)。NO与SoxR的[2Fe–2S]簇的反应几乎定量进行,同时还原消除了两个当量S 0原子。但是,中间的亚硝基化产物太不稳定而无法观察到。我们发现转化过程至少要经过两个步骤,其中较快的阶段是NO分子与[2Fe–2S]团簇的第一个反应。但是,与第二当量NO分子的反应较慢,对于DNIC的形成很重要。第三,为了阐明两种不同物种的SoxR蛋白不同反应之间的差异,我们使用诱变方法研究了大肠杆菌和铜绿假单胞菌SoxR与超氧阴离子的相互作用。尽管之间的同源性的大肠杆菌SoxR和铜绿假单胞菌SoxR,功能绿脓杆菌SoxR不同于的大肠杆菌。的取代大肠杆菌SoxR赖氨酸残基,位置靠近的[2Fe-2S]簇,到铜绿假单胞菌SoxR显着影响与超氧阴离子反应。











































 京公网安备 11010802027423号
京公网安备 11010802027423号