论文
2023
78. Jinzhe Zeng, Liqun Cao, Tong Zhu*, Neural network potentials, Quantum Chemistry in the Age of Machine Learning, 2023, 279.

2022
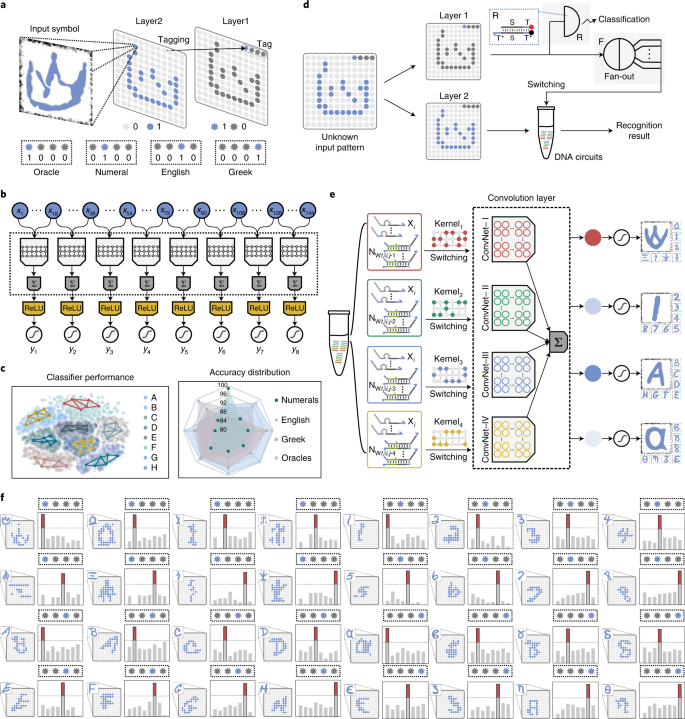
77. Xiewei Xiong+, Tong Zhu+, Yun Zhu, Mengyao Cao, Jin Xiao, Li Li, Fei Wang, Chunhai Fan, Hao Pei*, Molecular convolutional neural networks with DNA regulatory circuits, Nature Machine Intelligence, 2022, 4, 625.
76. Ya Gao, Bo Wang, Shiyu Hu, Tong Zhu*, John ZH Zhang*,An efficient method to predict protein thermostability in alanine mutation, Physical Chemistry Chemical Physics, 2022,24, 29629.
75. Liqun Cao, Jinzhe Zeng, Bo Wang, Tong Zhu*, John ZH Zhang*,Ab initio neural network MD simulation of thermal decomposition of a high energy material CL-20/TNT,Physical Chemistry Chemical Physics, 2022,24, 11801.
74. Xiaoniu Fang, Ya Gao*, Chen Wang, Huimin Chen, Tong Zhu*, Exploring the selective mechanism of inhibitors towards different subtypes of class I PI3K, Chemical Physics Letters, 2022, 786, 139174.
73. Chuchu Li, Yuqiao Han, Zhengyang Wang, Yanan Yu, Chen Wang, Ziwei Ren, Yanzhi Guo, Tong Zhu, XuWen Li*, Suzhen Dong*, Mingliang Ma*, Function-oriented synthesis of Imidazo[1,2-a]pyrazine and Imidazo[1,2-b]pyridazine derivatives as potent PI3K/mTOR dual inhibitors. European Journal of Medicinal Chemistry 2022, 247, 115030.
2021
72. Xu, M.; Zhu T.*; Zhang, J. Z.*, Automated Construction of Neural Network Potential Energy Surface: The Enhanced Self-Organizing Incremental Neural Network Deep Potential Method. J. Chem. Inf. Model 2021, 61, 5425.

71. Na Li, Ya Gao, Feng Qiu, Tong Zhu, Benchmark Force Fields for the Molecular Dynamic Simulation of G-Quadruplexes, Molecules 2021, 26, 5379.
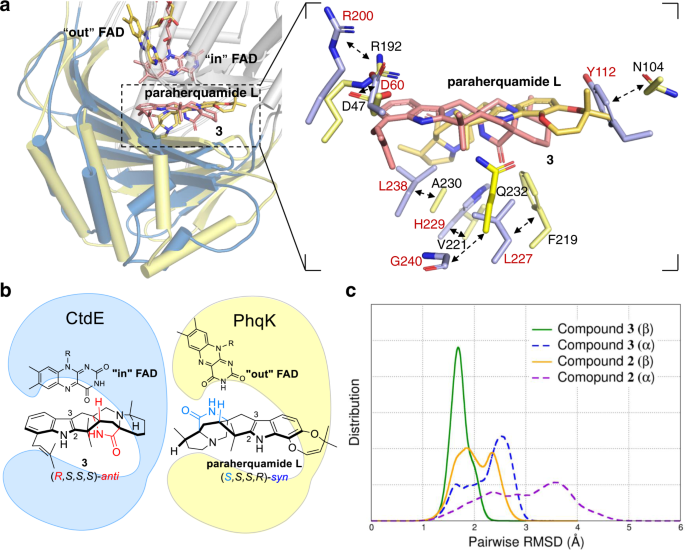
70. Zhiwen Liu, Fanglong Zhao, Boyang Zhao, Jie Yang, Joseph Ferrara, Banumathi Sankaran, BV Venkataram Prasad, Biki Bapi Kundu, George N Phillips, Yang Gao, Liya Hu, Tong Zhu.*, Xue Gao*, Structural basis of the stereoselective formation of the spirooxindole ring in the biosynthesis of citrinadins, Nature communications 2021,12, 4158.
69. Xu, M.; Zhu T.*; Zhang, J. Z.*, Automatically Construct Neural Network Potentials for Molecular Dynamics Simulation of Zinc Proteins. Frontiers in Chemistry 2021,9, 385.
68. Liu, Y.; Cong, Y.; Zhang, C.; Fang, B.; Pan, Y.; Li, Q.; You, C.; Gao, B.; Zhang, J. Z.; Zhu T.*, Zhang LJ*, Engineering the biomimetic cofactors of NMNH for cytochrome P450 BM3 based on binding conformation refinement. RSC Advances 2021,11 (20), 12036-12042.
67. Chin, C.-H.; Zhu, T.; Zhang, J. Z. H., Cyclopentadienyl radical formation from the reaction of excited nitrogen atoms with benzene: a theoretical study. Physical Chemistry Chemical Physics 2021,23 (21), 12408-12420.
66. Chen, J.; Li, N.; Wang, X.; Chen, J.*; Zhang, J. Z.; Zhu T.*, Molecular mechanism related to the binding of fluorophores to Mango-II revealed by multiple-replica molecular dynamics simulations. Physical Chemistry Chemical Physics 2021,23 (17), 10636-10649.
65. Cao, L.; Zeng, J.; Xu, M.; Chin, C.-H.; Zhu T.*, Zhang, J. Z.*, Fragment-Based Ab Initio Molecular Dynamics Simulation for Combustion. Molecules 2021,26 (11), 3120.
64. Zeng, J.; Zhang, L.*; Wang, H.*; Zhu T.* Exploring the Chemical Space of Linear Alkane Pyrolysis via Deep Potential GENerator. Energy & Fuels 2020.

2020
57. Z Gao, J Bao, S Shi, X Zhang, Y Gao, Zhu T.*,Effect of Mutations on Drug Resistance of Smoothened Receptor Toward Inhibitors Probed by Molecular Modeling. Chem. Phys. Lett. 2020, 741, 137126.
56. Xianli Hu, Bo Hou, Zhiai Xu, Madiha Saeed, Fang Sun, Zhenmei Gao, Yi Lai, Zhu T, Fan Zhang, Wen Zhang, Haijun Yu*.Supramolecular Prodrug Nanovectors for Active Tumor Targeting and Combination Immunotherapy of Colorectal Cancer. Adv. Sci. 2020, 7, 1903332
55. Zeng, J. Z.; Cao, L. Q.; Xu, M. Y.; Zhu T.*; Zhang, J. Z. H.*, Neural Network Based in Silico Simulation of Combustion Reactions. arXiv:1911.12252.
54. Zeng, J. Z.,‡ Cao, L. Q., ‡ Chin, C. H.;* Ren, H. S.;* Zhang, J. Z. H.; Zhu T.*, ReacNetGenerator: an automatic reaction network generator for reactive molecular dynamics simulations. Phys. Chem. Chem. Phys., 2020, 22, 683-691.
2019
53. Chin, C. H.*; Zhu T.*; Zhang J. Z. H., Formation mechanism and spectroscopy of C6H radicals in extreme environments: a theoretical study. Phys. Chem. Chem. Phys., 2019, 21, 23044-23055.
52. Tian, S. Z.; Zeng, J. Z.; Liu, X.*; Chen, J. Z.; Zhang, J. Z. H.; Zhu T.*, Understanding the selectivity of inhibitiors toward PI4KIIIa and PI4KIIIb based molecular modeling. Phys. Chem. Chem. Phys., 2019, 21, 22103-22112.
51. Chen, J. Z.*; Pang, L. X.; Wang, W.; Wang, L. F.; Zhang, J. Z. H.; Zhu T.*, Binding modes and conformational changes of FK506-binding protein 51 induced by inhibitor bindings: insight into molecular mechanisms based on multiple simulation technologies. J. Biomol. Struct. Dyn., 2019, 21, 22103-22112. J. Biomol. Struct. Dyn., 2019, DOI: 10.1080/07391102.2019.1624616.
50. Xu, M.Y.; Zhu T.*; Zhang J.Z.H.*, Molecular Dynamics Simulation of Zinc Ion in Water with an Ab Initio Based Neural Network Potential. J. Phys. Chem. A. 2019, 123, 6587-6595.
49. Tang, Q.; Plank, T. N.; Zhu T.; Yu H. Z.; Ge Z. L.; Li Q.; Li L.; Davis J. T.*; Pei H.*, Self-Assembly of Metallo-Nucleoside Hydrogels for Injectable Materials That Promote Wound Closure. ACS Appl. Mater. Interfaces. 2019, 11, 19743−19750
48. Zhao, P.; Han, S.; Li, X. X.*; Zhu T.*; Tao, X. F.; Guo, L., Comparison of RP-3 Pyrolysis Reactions Between Surrogates and 45-component Model by ReaxFF MD Simulations. ACS Energy Fuels. 2019, 33, 7176-7187.
47. Sun, Z. X.*; Wang, X. H.; Zhao, Q. Q.; Zhu, T., Understanding Aldose Reductase-Inhibitors interactions with free energy simulation. J. Mol. Graph. Model., 2019, 91, 10-21.
46. Chen, J. Z.*; Wang, X. Y.; Pang, L. X.; Zhang, J. Z. H.; Zhu T.*, Effect of mutations on binding of ligands to guanine riboswitch probed by free energy perturbation and molecular dynamics simulations. Nucleic Acids Res. 2019, 47, 6618-6631.
45. Zhao, P.; Cao, H. L.; Chen, Y.; Zhu T.*, Insights into the binding mechanisms of inhibitors of MDM2 based on molecular dynamics simulations and binding free energy calculations. Chem. Phys. Lett., 2019, 728, 94-101.
44. Chen, J. Z.*; Pang, L. X.; Wang, W.; Wang, L. F.; Zhang, J. Z. H.; Zhu T.*, Decoding molecular mechanism of inhibitor bindings to CDK2 using molecular dynamics simulations and binding free energy calculations. J. Biomol. Struct. Dyn., 2019, 8, 1-12.
43. Zhu, M. F.; Wang H.; Wang C. L.; Fang Y. F.; Zhu T.; Zhao W. L.; Dong X. C.*; Zhang X. W.*, L-4, a Well-Tolerated and Orally Active Inhibitor of Hedgehog Pathway, Exhibited Potent Anti-tumor Effects Against Medulloblastoma in vitro and in vivo. Front. Pharm. 2019, 10, 1-13.
42. Xu, M. Y.; Zhu T.*; Zhang, J. Z. H.*, A Fragment Quantum Mechanical Method for Metalloproteins. J. Chem. Theory Comput., 2019, 15, 1430–1439.
2018
41. Chen, J. Z.*; Wang, X. Y.; Zhang, J. Z. H.; Zhu T.*, Effect of Substituents in Different Positions of Aminothiazole Hinge-Binding Scaffolds on Inhibitor–CDK2 Association Probed by Interaction Entropy Method. ACS Omega, 2018, 3, 18052–18064.
40. Gao, Y.; Zhu T.*; Zhang, C. M.; Zhang, J. Z. H.; Mei, Y.*, Comparison of the Unfolding and Oligomerization of Human Prion Protein under acidic and neutral environments by Molecular Dynamics Simulations. Chem. Phys. Lett. 2018, 706, 594-600.
39. Gao, Y.; Zhu, T.*; Chen, J. Z.*, Exploring Drug-resistant Mechanisms of I84V Mutation in HIV-1 Protease toward Different Inhibitors by Thermodynamics Integration and Solvated Interaction Energy Method. Chem. Phys. Lett. 2018, 706, 400-408.
38. Yang, Q.; Hu, Z.; Zhu, S.; Ma, R.; Ma, H.; Ma, Z.; Wan, H.; Zhu, T.; Jiang, Z.; Liu, W.; Jiao, L.; Sun, H.; Liang, Y.; Dai, H., Donor Engineering for NIR-II Molecular Fluorophores with Enhanced Fluorescent Performance. J. Am. Chem. Soc. 2018, 140, 1715-1724.
37. Xu, M.; Zhu, T.*; Zhang, J. Z. H.*, A Force Balanced Fragmentation Method for ab Initio Molecular Dynamic Simulation of Protein. Front. Chem. 2018, 6, 189.
36. Wang, X.*; Li, Y.; Gao, Y.; Yang, Z.; Lu, C.; Zhu, T.*, A quantum mechanical computational method for modeling electrostatic and solvation effects of protein. Sci. Rep. 2018, 8, 5475.
35. Jin, X.; Zhu, T.; Zhang, J. Z. H.; He, X., Automated Fragmentation QM/MM Calculation of NMR Chemical Shifts for Protein-Ligand Complexes. Front. Chem. 2018, 6, 150.
2017
34. Xi, J.-B.; Fang, Y.-F.; Frett, B.; Zhu, M.-L.; Zhu, T.; Kong, Y.-N.; Guan, F.-J.; Zhao, Y.; Zhang, X.-W.; Li, H.-Y.; Ma, M.-L.; Hu, W., Structure-based design and synthesis of imidazo 1,2-a pyridine derivatives as novel and potent Nek2 inhibitors with in vitro and in vivo antitumor activities. Eur. J. Med. Chem. 2017, 126, 1083-1106.
33. Sun, Z.; Zhu, T.; Wang, X.; Mei, Y.; Zhang, J. Z. H., Optimization of convergence criteria for fragmentation methods. Chem. Phys. Lett. 2017, 687, 163-170.
32. Luo, Z.; Gao, Y.; Zhu, T.*; Zhang, J. Z.; Xia, F.*, Origins of Protons in C-H Bond Insertion Products of Phenols: Proton-Self-Sufficient Function via Water Molecules. J. Phys. Chem. A 2017, 121, 6523-6529.
31. Liu, L.; Zhao, F.; Liu, W.; Zhu, T.; Zhang, J. Z. H.; Chen, C.; Dai, Z.; Peng, H.; Huang, J.-L.; Hu, Q.; Bu, W.; Tian, Y., An Electrochemical Biosensor with Dual Signal Outputs: Toward Simultaneous Quantification of pH and O-2 in the Brain upon Ischemia and in a Tumor during Cancer Starvation Therapy. Angew. Chem. Int. Edit. 2017, 56, 10471-10475.
30. Li, S.; Zhu, A.; Zhu, T.; Zhang, J. Z. H.; Tian, Y., Single Biosensor for Simultaneous Quantification of Glucose and pH in a Rat Brain of Diabetic Model Using Both Current and Potential Outputs. Anal. Chem. 2017, 89, 6656-6662.
29. Jing, C.; Li, Z.; Jia, K.; Chen, C.; Liu, X.; Wang, B.; Hu, W.; Li, J.; Zhu, T*.; Dong, S.*, Discovery of Bisindole as a Novel Scaffold for Protein Tyrosine Phosphatase 1B Inhibitors. Arch. Pharm. 2017, 350, e1600173.
28. Huang, M.; Luo, Z.; Zhu, T.; Chen, J.; Zhang, J. Z.; Xia, F., A theoretical study of the substituent effect on reactions of amines, carbon dioxide and ethylene oxide catalyzed by binary ionic liquids. RSC Adv. 2017, 7, 51521-51527.
27. Gao, Y.; Zhang, C.; Wang, X.; Zhu, T.*, A test of AMBER force fields in predicting the secondary structure of alpha-helical and beta-hairpin peptides. Chem. Phys. Lett. 2017, 679, 112-118.
26. Duan, L. L.; Zhu, T.; Li, Y. C.; Zhang, Q. G.; Zhang, J. Z. H., Effect of polarization on HIV-1protease and fluoro-substituted inhibitors binding energies by large scale molecular dynamics simulations. Sci. Rep. 2017, 7, 42223.
25. Duan, L.; Zhu, T.; Ji, C.; Zhang, Q.; Zhang, J. Z. H., Direct folding simulation of helical proteins using an effective polarizable bond force field. Phys. Chem. Chem. Phys. 2017, 19, 15273-15284.
24. Dong, S.; Lei, Y.; Jia, S.; Gao, L.; Li, J.; Zhu, T.; Liu, S.; Hu, W., Discovery of core-structurally novel PTP1B inhibitors with specific selectivity containing oxindole-fused spirotetrahydrofurochroman by one-pot reaction. Bioorg. Med. Chem. Lett. 2017, 27, 1105-1108.
2016
23. Wang, Y.; Liu, J. F.; Zhu, T.; Zhang, L. J.; He, X.; Zhang, J. Z. H., Predicted PAR1 inhibitors from multiple computational methods. Chem. Phys. Lett. 2016, 659, 295-303.
22. Lv, F.; Chen, C.; Tang, Y.; Wei, J.; Zhu, T.*; Hu, W*., New peptide deformylase inhibitors design, synthesis and pharmacokinetic assessment. Bioorg. Med. Chem. Lett. 2016, 26, 3714-3718.
21. Liu, X.; Liu, J. F.; Zhu, T.; Zhang, L. J.; He, X.; Zhang, J. Z. H., PBSA_E: A PBSA-Based Free Energy Estimator for Protein-Ligand Binding Affinity. J. Chem. Inf. Model. 2016, 56, 854-861.
20. Liu, W.; Lu, Y.; Chai, X.; Liu, X.; Zhu, T.; Wu, X.; Fang, Y.; Liu, X.; Zhang, X.*, Antitumor activity of TY-011 against gastric cancer by inhibiting Aurora A, Aurora B and VEGFR2 kinases. J. Exp. Clin. Canc. Res. 2016, 35, 183.
19. Jin, X.; Zhu, T.; Zhang, J. Z. H.; He, X.*, A systematic study on RNA NMR chemical shift calculation based on the automated fragmentation QM/MM approach. RSC Adv. 2016, 6, 108590-108602.
18. Fang, Y.; Kong, Y.; Xi, J.; Zhu, M.; Zhu, T.; Jiang, T.; Hu, W.; Ma, M.*; Zhang, X.*, Preclinical activity of MBM-5 in gastrointestinal cancer by inhibiting NEK2 kinase activity. Oncotarget 2016, 7, 79313-79327.
2015
17. Zhu, T.; Zhang, J. Z. H.*; He, X.*, Quantum Calculation of Protein NMR Chemical Shifts Based on the Automated Fragmentation Method. In Advance in Structural Bioinformatics, Wei, D.; Xu, Q.; Zhao, T.; Dai, H., Eds. 2015; Vol. 827, pp 49-70.
16. Swails, J.; Zhu, T.; He, X.*; Case, D. A.*, AFNMR: automated fragmentation quantum mechanical calculation of NMR chemical shifts for biomolecules. J. Biomol. NMR 2015, 63, 125-139.
15. Liu, J.; Zhu, T*.; Wang, X.; He, X.*; Zhang, J. Z. H*., Quantum Fragment Based ab Initio Molecular Dynamics for Proteins. J. Chem. Theory Comput. 2015, 11, 5897-5905.
14. Chen, J.; Wang, X.; Zhu, T.*; Zhang, Q.; Zhang, J. Z. H., A Comparative Insight into Amprenavir Resistance of Mutations V32I, G48V, I50V, I54V, and I84V in HIV-1 Protease Based on Thermodynamic Integration and MM-PBSA Methods. J. Chem. Inf. Model. 2015, 55, 1903-1913.
2010-2014
13. Zhu, T.; Zhang, J. Z. H.; He, X.*, Correction of erroneously packed protein’s side chains in the NMR structure based on ab initio chemical shift calculations. Phys. Chem. Chem. Phys. 2014, 16, 18163-18169.
12. He, X.*; Zhu, T.; Wang, X.; Liu, J.; Zhang, J. Z. H.*, Fragment Quantum Mechanical Calculation of Proteins and Its Applications. Accounts Chem. Res. 2014, 47, 2748-2757.
11. Duan, L. L.; Zhu, T.; Zhang, Q. G.; Tang, B.*; Zhang, J. Z. H.*, Electronic polarization stabilizes tertiary structure prediction of HP-36. J. Mol. Model. 2014, 20.
10. Chen, J.; Chen, H.; Zhu, T.; Zhou, D.; Zhang, F.; Lao, X.*; Zheng, H.*, Asp120Asn mutation impairs the catalytic activity of NDM-1 metallo-beta-lactamase: experimental and computational study. Phys. Chem. Chem. Phys. 2014, 16, 6709-6716.
9. Zhu, T.; Zhang, J. Z. H.; He, X.*, Automated Fragmentation QM/MM Calculation of Amide Proton Chemical Shifts in Proteins with Explicit Solvent Model. J. Chem. Theory Comput. 2013, 9, 2104-2114.
8. Zhu, T.; Xiao, X.; Ji, C.*; Zhang, J. Z. H.*, A New Quantum Calibrated Force Field for Zinc-Protein Complex. J. Chem. Theory Comput. 2013, 9, 1788-1798.
7. Duan, L. L.; Zhu, T.; Mei, Y.*; Zhang, Q. G.; Tang, B.; Zhang, J. Z. H.*, An implementation of hydrophobic force in implicit solvent molecular dynamics simulation for packed proteins. J. Mol. Model. 2013, 19, 2605-2612.
6. Zhu, T.; He, X.*; Zhang, J. Z. H., Fragment density functional theory calculation of NMR chemical shifts for proteins with implicit solvation. Phys. Chem. Chem. Phys. 2012, 14, 7837-7845.
5. Zhu, T.; Hu, G. D.; Zhang, Q. G.*, Quasi-classical Trajectory Study of Reaction O (P-3) plus HCl (v=2; j=1, 6, 9) -> OH plus Cl. Chinese Phys. Lett. 2010, 27.
4. Zhu, T.; Hu, G.-D.; Chen, J. Z.; Liu, X. G.; Zhang, Q. G., Theoretical study of stereodynamics for reaction O(P-3)+HCl. Chinese Phys. B 2010, 19.
3. Zhu, T.; Hu, G.; Zhang, Q.*, Quasi-classical trajectory study of the reaction O(P-3) plus HCl -> OH + Cl and O(P-3) plus DCl -> OD plus Cl: Vector and scalar properties. J. Mol. Struc-Theochem 2010, 948, 36-42.
2. Liu, H.; Liu, X.; Zhu, T.; Sun, H.; Zhang, Q.*, THE EFFECT OF VIBRATIONAL EXCITATION OF THE REACTION O(P-3)+HCl -> OH plus Cl FOR THE (3)A ” ELECTRONIC STATES. J. Theor. Comput. Chem. 2010, 9, 1033-1042.
1. Hu, G.-D.; Zhu, T.; Zhang, S.-L.; Wang, D.; Zhang, Q.-G.*, Some insights into mechanism for binding and drug resistance of wild type and I50V V82A and I84V mutations in HIV-1 protease with GRL-98065 inhibitor from molecular dynamic simulations. Eur. J. Med. Chem. 2010, 45, 227-235.50