Abstract
Mycobacterium bovis is the causative agent of bovine tuberculosis and the predominant cause of zoonotic tuberculosis in people. Bovine tuberculosis occurs in farmed cattle but also in a variety of wild animals, which form a reservoir of infection. Although direct transmission of tuberculosis occurs between mammals, the low frequency of contact between different host species and abundant shedding of bacilli by infected animals suggests an infectious route via environmental contamination. Other intracellular pathogens that transmit via the environment deploy strategies to survive or exploit predation by environmental amoebae. To explore if M. bovis has this capability, we investigated its interactions with the soil and dung-dwelling amoeba, Dictyostelium discoideum. We demonstrated that M. bovis evades phagocytosis and destruction by D. discoideum and actively transits through the amoeba using the ESX-1 Type VII Secretion System as part of a programme of mechanisms, many of which have been co-opted as virulence factors in the mammalian host. This capacity of M. bovis to utilise an environmental stage between mammalian hosts may enhance its transmissibility. In addition, our data provide molecular evidence to support an evolutionary role for amoebae as training grounds for the pathogenic M. tuberculosis complex.
Similar content being viewed by others
Introduction
Mycobacterium bovis, a member of the M. tuberculosis complex, causes bovine tuberculosis, one of the most important veterinary health problems in the UK and Ireland [1, 2]. It is also a neglected zoonotic infection in humans, causing an estimated 147,000 new infections and 12,500 deaths worldwide in 2016 [3, 4]. In addition to farmed cattle, a wide range of wild mammals harbours the disease including badgers (UK), wild boar and red deer (continental Europe) and brushtail possums (New Zealand), and likely act as endemic wildlife reservoirs for the disease [5, 6]. Transmission from a wildlife reservoir in the UK and Ireland is thought to be a prominent factor undermining the effectiveness of control measures based on cattle test and slaughter. Transmission of bovine TB is primarily mediated through inhalation of aerosols from infected animals during close contact [7]. However, the exact means of transmission to and from the wildlife reservoir remains to be fully elucidated. For example, tracking of cattle and badgers has shown that they do not come into close contact on pasture [8]. However, badgers and cattle have been shown to secrete bacilli in their urine and/or faeces, and M. bovis has been detected in the soil and pasture near badger setts [9, 10]. This, and the finding that M. bovis can survive for extended periods in soil [11], has led researchers to postulate that indirect transmission through environmental contamination may facilitate the spread of bovine tuberculosis [1, 2, 12]. Although M. tuberculosis complex organisms are host-adapted species, their evolutionary origins lie with environmental organisms [13, 14]. As such, both M. bovis and its non-pathogenic progenitors are likely to have interacted with unicellular eukaryotes, such as species of amoeba that are abundant in terrestrial or aquatic environments.
It is postulated that contact with amoebae may have shaped the evolution of mycobacteria [15,16,17,18] with the amoeba intracellular environment acting as an evolutionary “training ground” for pathogenic bacteria [19,20,21,22,23]. Studies on Mycobacterium marinum, a pathogen of poikilotherms such as fish and frogs [14, 24,25,26] and the environmental opportunistic pathogens of the M. avium complex [27, 28], support this view, with overlap in the virulence mechanisms/genes used to replicate and survive in both amoebae and the phagocytes of higher organisms. In M. marinum, this includes the involvement of the ESX-1 Type VII Secretion System to effect phagosome escape [17, 29,30,31].
Given the potential importance of an environmental element to the transmission cycle of M. bovis between hosts, we surmised that it would be advantageous for M. bovis to possess active survival mechanisms to endure predation by environmental amoebae. To investigate this, we examined the uptake, survival and escape of this mammalian pathogen upon interaction with the free-living soil and dung-dwelling amoeba, Dictyostelium discoideum [32]. Further, we screened M. bovis genetic mutants to reveal that escape from the host amoeba required numerous genes/molecular mechanisms that are also required for full virulence of M. tuberculosis complex bacteria in mammals. This is supportive of an evolutionary history in which adaptation to intracellular survival first occurred in amoebae and was later co-opted by the M. tuberculosis complex and other pathogenic mycobacteria for infection of phagocytes from vertebrates.
Materials and methods
Bacterial strains and culture methods
Mycobacterium bovis AF2212/97 [33], Genbank Accession NC_00000962, was maintained on Middlebrook 7H11 solid medium containing 40 mM sodium pyruvate, 0.5% lysed defibrinated sheep blood, 5% heat inactivated foetal bovine serum and 10% oleic acid-albumin-dextrose-catalase (OADC) supplement. M. bovis was cultured as a liquid broth in 7H9 medium containing 40 mM sodium pyruvate, 0.05% Tween®80 and 10% OADC. M. marinum M strain, M. bovis BCG and M. smegmatis mc2155 were maintained on 7H11 solid medium supplemented with 0.5% glycerol and 10% OADC, and in liquid broths using 7H9 medium supplemented with 0.05% Tween®80, 0.2% glycerol and 10% OADC.
Construction of a ΔespAC mutant strain of M. bovis
A double knockout M. bovis strain of espA (Mb3646c; Rv3616c) and espC (Mb3645c; Rv3615c) was made using plasmids and primers as previously reported [34]. Genomic sequence upstream (left flank) and downstream (right flank) of Mb3645c-3646c were PCR amplified and cloned either side of the zeocin antibiotic resistance cassette of the cosmid pANE001 (pYUB854) [35] with the hygromycin cassette replaced with a zeocin cassette. Transduction phage were constructed and transduced into M. bovis using the pHAE159 mycobacteriophage-based method of transduction [35], and recovered on 7H11 with 25 μg/ml zeocin. Knockouts were confirmed by PCR using primers outside of the upstream and downstream flanking regions both alone and in combination with antibiotic cassette specific primers.
Culture/Infection of D. discoideum with bacteria
The axenic D. discoideum laboratory strain AX2 was maintained at 20–80% confluence in 10-cm tissue culture dishes (Falcon) in HL5c medium including glucose and supplemented with vitamins and microelements (Formedium, Hunstanton, UK). For maintenance, penicillin (100 U/ml) and streptomycin (100 µg/ml) were included in the culture medium. For cultures with bacteria, D. discoideum were seeded into dishes in medium without penicillin and streptomycin.
For bacterial survival assays in D. discoideum, M. bovis, M. marinum and M. smegmatis were prepared for infection by harvesting at mid-late log phase (OD 0.8–1.0), centrifuging at 3700 × g for 20 min and re-suspending in HL5c medium without antibiotics. Confluent monolayers of 2 × 107 were gravity infected with an MOI of 10 bacilli per amoeba for 2.5 h. The supernatant was removed and the monolayer of amoebae was washed extensively with HL5c medium to remove extracellular bacilli. D. discoideum were harvested by gently scraping, diluted and seeded in tissue culture flasks. Where indicated, 5 μg/ml streptomycin was added to the culture medium to prevent extracellular proliferation. D. discoideum were harvested by gently scraping and by centrifugation at 1500 × g for 10 min. D. discoideum were lysed using 0.1% Triton X-100 in water, and the bacilli released were recovered onto 7H11 medium. For differential separation of D. discoideum-associated and extracellular bacilli, the D. discoideum fraction was separated by centrifugation at 250 × g for 10 min, before recovery of bacilli by centrifugation at 3700 × g for 30 min.
For bacterial uptake assays, D. discoideum were cultured with FITC-labelled K12 E. coli (Life Technologies, Carlsbad, CA, USA), and FITC-labelled M. bovis BCG [36] for 2 h before washing in HL5c medium to remove extracellular bacteria, gently scraping to a suspension, washing with Sorensen’s phosphate buffer containing 5 mM sodium azide, and re-suspending in Sorensen’s phosphate buffer containing 120 mM sorbitol. Phagocytosis was determined by flow cytometry using an Attune Flow Cytometer (Life Technologies, Carlsbad, CA, USA), using the gating strategy described by Hagedorn and Soldati [37]. To assess internalisation by confocal microscopy, D. discoideum were seeded in 35 mm glass-bottomed imaging dishes (ibidi, Graefelfing, Germany) and infected with FITC-labelled M. bovis BCG as before. External bacteria were gently rinsed away and the amoebae fixed for 3 h at 4 °C with 4% methanol-free paraformaldehyde in Sorensen’s phosphate buffer. Amoebae were rinsed with Sorensen’s phosphate buffer and permeablised for 3 min at room temperature with PBS containing 0.1% Triton X-100. Actin was stained with ActinRed555 ReadyProbes reagent and nuclei visualised with NucBlue Fixed Cell ReadyProbes reagent (both from Molecular Probes). Amoebae were rinsed again and mounted with a coverslip and Prolong Gold (Molecular Probes). Amoebae were visualised using a Nikon A1M Confocal Microscope, using a ×60 oil immersion lens, and sequential scanning using the 405 nm, 488 nm and 561 nm laser lines. For assessing the total DNA content of cultures, M. bovis was fixed for 24 h with 4% paraformaldehyde, washed with PBS, and DNA labelled with SYTOX Green nucleic acid stain (Molecular Probes). Fluorescence measurements were performed using an Ascent Fluorescent plate-reader in black walled clear bottomed plates using the FITC filter sets.
M.bovis transposon mutant library construction and selection for mutants trapped in D. discoideum
A transposon mutant library of M. bovis AF2212/97 was generated according to Long et al. [38]. Briefly, 100 ml of late log M. bovis culture was washed twice in MP buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgSO4 and 2 mM CaCl2) at 37 °C, and then incubated with more than 1 × 1011 pfu of φMycoMarT7 phage for 4.5 h. The suspension was recovered by centrifugation, washed and plated on ten 15-cm 7H11 plates supplemented with 20 μg/ml kanamycin and 0.02% Tween®80. After 18 days the library was scraped from the plates, dispersed in broth overnight and frozen in aliquots at −80 °C in 10% glycerol. For library selection, M. bovis-Tn library was thawed and washed in 7H9 before recovering in 7H9 complete medium containing 30 μg/ml kanamycin at 37 °C for 4 days. Mid log (OD 600 0.4–0.6) bacilli were prepared as before for infection, and used to infect 4 × 107 D. discoideum at an MOI of 10 for 3 h in tissue culture coated dishes. In addition, 2 × 107 cfu of library was plated onto 5 × 15 cm dishes of 7H11 containing 30 μg/ml kanamycin as the library reference control for each experiment. After 3 h, D. discoideum were washed with HL5c medium to remove extracellular bacilli, gently scraped and diluted into large tissue culture flasks. Streptomycin was omitted from these experiments as HL5c medium does not support the in vitro replication of M. bovis (Supplementary Fig. 1). After 2 days, D. discoideum were gently scraped and separated from extracellular bacteria by differential centrifugation. Bacilli were released from D. discoideum by lysing as before, and plating onto 5 × 15 cm dishes of 7H11 supplemented with 30 μg/ml kanamycin. After 3–4 weeks, recovered M. bovis was harvested and stored in 10% glycerol at −80 °C. Four independent co-culture/infections were performed, three of which included technical duplicates that were harvested and sequenced independently.
TnSeq: Genomic DNA preparation and transposon-insertion site sequencing
M. bovis mutant pools were resuspended in TE buffer pH 8.0, and incubated for 5 min rocking with an equal volume of methanol:chloroform 2:1. Bacilli were centrifuged and the pellet dried, before re-suspending in phenol:chloroform:isoamyl alcohol 25:24:1. Cells were disrupted with the Fastprep homogeniser (MP Biomedicals, Irvine, CA, USA) and lysing matrix B, and centrifuged to separate the aqueous and organic phases. The upper aqueous phase was re-extracted with an equal volume of phenol:chloroform:isoamyl alcohol 25:24:1, followed by 2 further extractions with chloroform. gDNA was precipitated with 0.1 volume 3 M sodium acetate pH 5.2 and 1 volume propan-2-ol, pelleted by centrifugation, washed with 70% ethanol, and resuspended in TE. Transposon junctions were amplified essentially as described in [34]. Briefly, a 5 μg aliquot of DNA was sheared in a Covaris® Sono 7 machine (Covaris Inc, Woburn, MA, USA) and purified using SPRIselect beads (Beckman Coulter, High Wycombe, UK) at 1× concentration. The DNA fragment ends were repaired, blunt ended, and ‘A’ overhangs added, using the NEBNextEnd repair/dA-Tailing Module protocol (NEB, Hitchin, UK). Linkers (annealed Adap1 and Adap2, Supplementary Table 1) were ligated to the ‘A’ tail ends at ×100 molar excess with the NEBNext Ultra II Ligation module (NEB) and the fragments purified with SPRIselect as before. Transposon junctions were amplified using primers IS6 and MarA to MarO (Supplementary Table 1), cycle conditions were 95 °C for 10 s, 58 °C for 10 s, 72 °C for 30 s. The primers have a P5-index that identifies the sample and a random P7-index to allow PCR-generated artefacts to be identified and removed from the data. Real-time PCR was used to determine the minimum number of cycles required to amplify the transposon junctions. PCR products were purified with SPRIselect and equimolar quantities of each preparation sequenced using a HiSeq® 2500 (Illumina, San Diego, CA, USA) with a single sequence read and double index reads.
Analysis of transposon site data
Sequence data were demultiplexed using the P5-index reads, and quality controlled and aligned as described [39]. An additional step [34] removed artefactual amplicons generated by the PCR amplifications, which were identified as those reads having both identical P7-indexed reads and the same insertion site. Reads were aligned to the M. bovis AF2212/97 genome (NC_00000962) using Bowtie and the transposon counts determined. Gene essentiality in the input library was predicted using the TRANSIT Hidden Markov model (HMM) method [40] but excluding TA sites with a non-permissive motif as identified previously in M. tuberculosis [41]. We determined the mutants that were significantly enriched in the intracellular Dictyostelium fraction by comparing mutant abundance (numbers of each unique TnSeq read per gene) in the Dictyostelium-trapped fraction and the inoculating mutant library using the resampling option in the TRANSIT software with nzMean normalisations and non-permissive sites removed. This test uses a variation of the classical permutation statistical test, to provide a p value, which is adjusted for multiple comparisons by the Benjamini Hochberg procedure to provide a q value. Mutants were considered to be enriched or depleted in a condition at q < 0.05. The significance of gene group/pathway enrichments were determined using Fisher’s exact test
Results
Mycobacterium bovis survives predation by Dictyostelium discoideum amoeba
D. discoideum is a bacterivorous amoeba that survives in soil by preying on bacteria. To assess whether M. bovis could be a predated food source for D. discoideum, we compared the efficiency of uptake of FITC-labelled M. bovis BCG and E. coli K12 by flow cytometry. As seen in Fig. 1a, M. bovis BCG is poorly phagocytosed by D. discoideum in comparison with E. coli K12. However, internalisation of M. bovis BCG was confirmed by confocal microscopy (Fig. 1b). We next investigated the survival of M. bovis AF2122/97 in D. discoideum over 2 days of infection, and compared it with the survival of the pathogenic mycobacterial species M. marinum and non-pathogenic M. smegmatis. As seen in Fig. 1c–e, and in agreement with Hagedorn et al. [37], the pathogenic mycobacterial species M. marinum survives a 2-day infection in D. discoideum, whereas non-pathogenic M. smegmatis is killed by amoeba. We demonstrate that M. bovis also survives for 2 days in D. discoideum. Taken together, our data demonstrate that M. bovis is inefficiently phagocytosed as prey by D. discoideum; however, once phagocytosed, M. bovis is resistant to intracellular killing.
a The percentage of D. discoideum that have phagocytosed E. coli K12-FITC or M. bovis BCG-FITC after a 2 h incubation at an MOI of 10 was determined by flow cytometry. The mean of three biological replicates ± SEM is displayed. b Representative micrograph of D. discoideum demonstrating internalised M. bovis BCG-FITC (green—M. bovis BCG-FITC; red—actin; blue—nuclei). D. discoideum was infected with an MOI of 10 with (c) M. marinum M strain, (d) M. smegmatis mc2155 strain, or (e) M. bovis AF2122/97. Cfu of bacteria per culture of 2 × 107 amoebae was enumerated at the point of infection and after 2 days to determine bacterial survival. Streptomycin was included in the medium post infection to inhibit extracellular replication of bacteria. The mean and SEM of n = 3 independent experiments is displayed.
Mycobacterium bovis replicates at ambient temperature
D. discoideum is unable to tolerate the high temperatures of warm-blooded animals, with an upper temperature growth limit of around 25.5 °C. The environmental species M. marinum has an overlapping temperature range and thus is able to divide in D. discoideum at 25 °C. However, as a pathogen of mammals, M. bovis replicates in cattle with a body temperature of 38.5 °C, and as such might not be expected to efficiently replicate at lower temperatures. We compared the growth of M. bovis in optimal growth medium at 25 and 37 °C. As seen in Fig. 2, over 14 days of growth the optical density of the cultures increases, indicating slow but detectable bacterial division at 25 °C (Fig. 2a). By using a dye to measure total DNA content in the culture, we confirmed that the increase in culture density was indeed due to bacterial division rather than an artefact caused by the disaggregation of bacterial clumps (Fig. 2b). Counting cfu revealed that there was an apparent balance of division and progression to a non-culturable state in the 25 °C cultures, such that the viable plate count did not significantly increase over the 14-day time period analysed (Fig. 2c). Taken together, our data demonstrate that M. bovis is metabolically active and divides at a much lower temperature than previously reported. This observation is important because it allows for the possibility that M. bovis survival in D. discoideum is an active process. Furthermore, survival/adaptation to ambient temperatures may partially explain the success of M. bovis transmission between cattle, and between cattle and wildlife reservoirs, which has been postulated to occur via an intermediate environmental step.
a Growth of M. bovis AF2122/97 was compared at 25 and 37 °C by optical density (OD600 nm) at 0 (n = 4), 4 (n = 3), 7 (n = 4) and 14 (n = 2) days. The mean and SEM of independent experiments (each performed in biological duplicate or triplicate) is displayed. Growth was compared with day 0 by two-way ANOVA (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001). b DNA synthesis in cultures was measured using Sytox Green fluorescent dye, (mean of three biological replicates ± SEM is displayed). c Growth of M. bovis AF2122/97 was compared at 25 and 37 °C by counting colony forming units. The mean and SEM of n = 2 independent experiments (each performed in biological triplicate) is displayed. Growth was compared with day 0 by two-way ANOVA (**p < 0.01).
Mycobacterium bovis uses the ESX-1 secretion system to escape from D. discoideum
The ESX-1 locus encodes a Type VII Secretion System that is required for virulence in M. bovis and M. tuberculosis in mammals [30, 42,43,44,45]. Mycobacteria with mutations in the ESX-1 machinery have been shown to have reduced fitness in mammalian phagocytes, mice and guinea pigs [39, 46,47,48]. The ESX-1 machinery transports virulence factors into host cells, including the immunodominant co-secreted effectors, EsxA (ESAT-6) and EsxB (CFP-10). EsxA has the ability to disrupt cell membranes [49, 50], potentially catalysing the ESX-1-dependent exit of M. tuberculosis complex bacteria from the phagosome of mammalian cells into the cytosol [51,52,53,54,55] and ultimately inducing host cell lysis and bacterial release [50, 56,57,58,59]. To determine if ESX-1 plays a role in M. bovis survival in amoebae, we compared the ability of wild type and ESX-1 defective M. bovis to transit through D. discoideum. We constructed a mutant of the espACD operon, which is essential for ESX-1-dependent EsxA/EsxB secretion and pathogenic function [60,61,62]. D. discoideum were allowed to ingest wild-type M. bovis and M. bovis ΔespAC, and, after 48 h, Dictyostelium-associated bacilli and extracellular bacilli were separated by differential centrifugation. As seen in Fig. 3, over 60% of wild-type M. bovis escaped from D. discoideum but loss of ESX-1 function by deletion of espAC impaired M. bovis escape. This contrasts with M. marinum where the loss of ESX-1 alters the route of bacterial release, reducing cytolysis but actually increasing transit [63]. There was no difference in the cytotoxic capacity of M.bovis wild-type and ΔespAC strains (Supplementary Fig. 2). In conclusion, M. bovis actively transits though D. discoideum, in part using the ESX-1 locus.
D. discoideum were infected at MOI 10 with M. bovis wild-type (WT) and M. bovis ΔespAC. After 3 h and 48 h, D. discoideum and extracellular bacteria were separated by differential centrifugation, and bacilli enumerated after recovery on solid medium. The experiments were performed in the absence of streptomycin; the HL5c growth medium does not support the extracellular replication of M. bovis (Fig. S2). The mean and SEM of n = 2 independent experiments (each performed in biological duplicate or triplicate) is displayed. Statistical analysis was performed by Student’s t test (*p < 0.05).
M. bovis genes associated with virulence in vertebrates are required for escape from D. discoideum
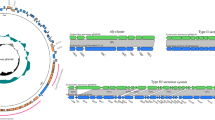
To further understand the involvement of ESX-1 in M. bovis escape from D. discoideum and to investigate other genes/systems involved in this interaction, we performed a genome-wide screen of M. bovis transposon-insertion mutants, selecting for those which had an impaired ability to transit through D. discoideum. A pool of ~3.5 × 105 individual mutants was generated and sequencing revealed that it contained ~43,000 unique transposon-insertion sites and 318 genes were categorised to be essential by analysis with TRANSIT HMM (Fig. 4a, Supplementary Table 2). D. discoideum was infected with the mutant library, and after 48 h the transposon mutants that remained trapped inside the amoebae were recovered by differential centrifugation and lysis of D. discoideum. This provided a positive selection for mutants with defective transit through the amoebae. M. bovis mutants from the D. discoideum-associated fractions of four independent experimental infections were recovered on 7H11 medium and the relative abundance of mutants in each pool compared by TnSeq with similarly recovered mutant pools of the inocula [40]. Mutations in 694 genes were significantly enriched in the intracellular D. discoideum fraction (Supplementary Table 2) indicative of defective transit through D. discoideum.
a The distribution of transposon-insertion sites around the bacterial chromosome that result in defective transit through D. discoideum are depicted in the outer ring (in blue). The line length represents the fold change after 2-day infection in D. discoideum, compared with the inoculum. The distribution of essential growth genes is represented by the inner ring (rust). b The distribution of mutations associated with defective transit through D. discoideum is displayed by functional class (outer ring). The distribution of genes containing transposon insertions in the unselected inoculum library is represented by the inner circle.
Genes required for transit through D. discoideum were distributed widely throughout the mycobacterial chromosome, including in the chromosomal regions that are overtly devoid of essential growth genes (Fig. 4a). We analysed the distribution of these mutations within functional categories of genes assigned by Mycobrowser (Fig. 4b). Mutations associated with impaired escape from D. discoideum are distributed throughout all functional classes, reminiscent of virulence gene datasets for M.tuberculosis in vertebrates [47, 64]. We further analysed the amoeba-trapped mutations to assess the statistical significance of the “escape” phenotype with known gene families, pathways and functional groups in M. bovis (Table 1).
ESX-1 secretion system
Consistent with findings that the ESX-1 machinery is required for transit of M. bovis through D. discoideum, we found that mutations in 12 genes of the extended ESX-1 locus (ESX-1 Mb3894-3913, plus the espACD operon and EspR regulator) are enriched in the intracellular D. discoideum mutant pool (Supplementary Table 2, Table 1 and Fig. 5). By contrast, mutations in the related type VII secretion systems ESX-2, ESX-3, ESX-4 and ESX-5 were not significantly associated with impaired transit through amoebae. However, as ESX-3 is an essential system and mutations in ESX-5 cause an in vitro growth defect (Supplementary Fig. 3), definitive conclusions about their specific involvement in the bacterium-amoeba interaction cannot be drawn.
A model for ESX-1 secretion (adapted from [89,90,91]). EsxA/EsxB and EspA/EspC bind to cytoplasmic ATPases, such as EccA1 and EccCb1, via the C-terminals of EsxB and EspC, respectively. The protein complex interacts with the translocation complex likely formed by the transmembrane proteins EccCa1, EccB1, EccD1, EccE1 and MycP1 [90]. When uncoupled from the chaperone protein EspA, EspC is able to polymerise into filaments which may form the secretion system across the mycomembrane [92].
PE/PPE proteins
The PE/PPE proteins are unique to mycobacteria and have been previously associated with virulence in vertebrates. Here, mutants carrying insertions in PE/PPE family proteins were prominently enriched in D. discoideum (Fig. 4b, dark grey segments, Table 1) with mutation of 27 PE and 24 PPE genes conferring attenuation for amoeba transit (Supplementary Table 2). Further analysis of the PE family revealed that the mutants of 20 PE-PGRS proteins were defective for amoeba escape.
MCE4 lipid transporter and lipases
Mutants in genes encoding the virulence-associated MCE4 cholesterol transport system [65] were also indicated as trapped in D. discoideum infection (Table 1, Supplementary Table 2, Supplementary Fig. 4). Export of the MCE4 machinery to the cell envelope has recently been shown to require the function of the SecA2 secretion apparatus [66]; we also find that the mutations in SecA2 are significantly enriched in our D. discoideum screen (Supplementary Table 2). Interestingly, mutations in Lip-family lipases (24 lipid/ester hydrolases called LipC to LipZ) were additionally found to be enriched in D. discoideum (Table 1, Supplementary Table 2), suggesting that utilisation of triacylglycerol-derived carbon may be required for optimal escape from amoeba.
Cell wall lipid synthesis
M. tuberculosis complex mutants lacking cell wall phthiocerol dimycocerosates (PDIM) are heavily attenuated in mammalian phagocytes [67] and act in concert with EsxA to disrupt phagosomal membranes [49, 68, 69] suggesting a mechanism by which the regulation of PDIM content of the cell envelope could aid escape from amoebae. However, although insertional mutations in the Mmpl7 PDIM transporter, and the key PDIM biosynthetic enzyme FadD26 [70] were found to be significantly enriched in D. discoideum (Supplementary Table 2), as a collective, insertional mutations in PDIM synthetic pathways were not significantly enriched. By contrast, mutations in genes encoding for the synthesis and transport of sulfolipid-1 (SL-1), a virulence-associated outer membrane glycolipid [71] were attenuated for D. discoideum escape (Table 1, Supplementary Table 2, Supplementary Fig. 4).
Other vertebrate-virulence/Dictyostelium-escape genes
Other parallels with M. tuberculosis virulence genes that may shed some light on the dynamic nature of the D. discoideum/M. bovis interaction are the importance of genes involved in the oxidative stress response (oxyS ROS-responsive regulator and the katG catalase gene) and genes for uptake of aspartate, ansP1, and asparagine, ansP2. [72] (Supplementary Table 2). Evidence that M. bovis needs to actively sense the intra-amoebic environment in order to respond and exit the amoeba host cell is supported by attenuation of a significant number of mutants in serine/threonine protein kinases (STPKs, Table 1 and Supplementary Table 2).
Discussion
This study demonstrates for the first time that a member of the M. tuberculosis complex is adapted to actively combat predation/phagocytosis by amoebae at environmental temperature. This observation is important at two levels: understanding the transmission of M. bovis between animal hosts; and, understanding the evolution of pathogenesis in the M. tuberculosis complex. Whereas human tuberculosis is spread by aerosol infection directly between hosts in close contact, the transmission route of bovine tuberculosis between cattle and badgers remains to be fully elucidated. The secretion of M. bovis in urine and faeces of infected animals and the finding that M. bovis can survive for extended periods in soil, has led researchers to postulate that environmental contamination may facilitate the spread of bovine tuberculosis [1]. For this transmission route to be effective, we hypothesised that M. bovis possessed adaptations for environmental survival including mechanisms that allow it to survive predation by bacterivorous amoebae that reside in soil and dung [32], like D. discoideum.
Arguably the most significant observation of the present study was that M. bovis displayed active metabolism and cell division at 25 °C, thus it has the physiological opportunity to adapt to the environment, including amoebae, between vertebrate hosts. The role of amoebae in the transmission cycle of M. bovis has been a source of debate. One school of thought is that amoebae may provide a protective or replicative niche for the mycobacterium. Indeed, M. bovis has been shown to survive within the trophozoites of Acanthamoeba polyphaga and A. castellanii, [18, 73, 74] where a proportion become encased in a protective cyst and maintain the ability to cause infection in mice [73]. However, the majority of M. bovis bacilli bypass the amoeba cyst stage, possibly by exiting the protozoan via exocytosis or non-lytic ejection described in D. discoideum [17, 18, 74]. Consistent with this, and coherent with our hypothesis, we show that the bacterium is not phagocytosed in large quantities by D. discoideum but when it is internalised it uses mechanisms encoded by a large set of genes to effect escape from the amoeba.
Intriguingly, many of the factors required for escape from D. discoideum are previously documented as virulence factors in mammalian hosts, most conspicuously the ESX-1 system. ESX-1 in M. marinum is crucial for parasitism of amoebae, mammalian phagocytes, and zebrafish models of infection [17, 29, 31]. However, our demonstration that the mammalian pathogen M. bovis also uses ESX-1 to evade the microbicidal mechanisms of amoebae establishes more firmly that amoebae could have acted as evolutionary “training grounds” for the ancestors of today’s M. tuberculosis complex. Supporting this conclusion is the importance of PE/PPE proteins to both the escape of M. bovis from amoebae and in subversion of vertebrate immunity and control of macrophage function [75, 76]. PE/PPE protein families are unique to mycobacteria, and their expansion in the M. tuberculosis complex, where they account for ~10% of the coding capacity of the genome [77] has been previously linked to adaptation to human and animal hosts [71, 78]. This is clearly illustrated in the most recently evolved group of PE proteins, the PE-PGRS sublineage, which are exclusively associated with the M. tuberculosis complex and related pathogenic mycobacteria such as M. marinum [76, 78]. The present study identified 20 PE-PGRS genes important for amoeba escape, twelve of which were previously identified as important for M. tuberculosis survival in human dendritic cells [39]. This is of particular interest given that PE-PGRS are not secreted in M. bovis due to RD5-deletion of ppe38, suggesting that the virulence phenotype of these genes is not dependent on secretion [79]. It is tempting to speculate that the genomic expansion of the extensive PE/PPE gene/protein families may have been driven by selective pressure from amoebae and the adaptation co-opted for survival in vertebrate phagocytes.
We have identified other known “mammalian virulence” gene families involved in escape from D. discoideum, including those encoding the MCE4 cholesterol transport pathway. MCE4 has been implicated in transport of lipids across the mycobacterial cell wall including uptake of cholesterol [65] and it is important for the maintenance of infection in the murine model of TB [47, 80]. Although D. discoideum does not synthesise cholesterol, instead synthesising sitosterol [81], our results are consistent with those in M. marinum where MCE4 orthologues are required for growth in D. discoideum suggesting a dietary switch to sterols as a conserved adaptation for intracellular mycobacteria in phagocytes [31]. We also identify cell wall SL-1, to be required for D. discoideum phagosome escape. SL-1 is a glycoprotein which is only found in pathogenic mycobacteria [82], and has previously been implicated in survival in human monocyte-derived dendritic cells [39]. This is particularly intriguing because M. bovis possesses only small amounts of SL-1 sulfolipid in its cell envelope compared with M. tuberculosis [83], due to mutation of the PhoP/R two-component regulatory system [84], and had thus been previously overlooked as a molecule of importance to M. bovis biology.
We found that mutants in several serine threonine protein kinases including PknH were impaired in their ability to escape from D. discoideum (Table 1 and S1). PknH phosphorylates the regulator protein EmbR in M. tuberculosis, which regulates arabinogalactan synthesis via the embABC genes, and may control the balance of lipoarabinomannan and lipomannan in the cell envelope [85]. M. bovis contains a second regulator EmbR2, which is thought to negatively regulate EmbR [86]. Mutations in both EmbR and Embr2 were also found to be enriched in D. discoideum. Collectively this suggests that regulating the arabinogalactan content of the cell wall may be required for optimal escape from amoeba. Escape defective M.bovis mutants also included those deficient in previously described virulence determinants involved in resistance to oxidative stress and nutrient acquisition. Thus, there appears to be considerable depth in the intracellular “virulence” adaptations co-opted between amoebae and vertebrate phagocytes.
One potential limitation of the assay is that mutants with profound intra-dictyostelium mortality may not be identified by the positive selection strategy for trapped-intracellular bacteria. However, our preliminary experiments with M. bovis ΔespAC suggested that transit-defective bacteria are not necessarily killed during the relatively short 48 h co-culture experiment. Thus we are confident that the vast majority of mutants with an altered intra-amoeba phenotype will be identified by the experiment.
We hope that our observations on D. discoideum/M. bovis interactions will encourage researchers to utilise D.discoideum as a useful, non-sentient, genetically tractable model of tuberculosis with the potential to reveal aspects of mycobacterial pathogenesis from an alternative but biologically relevant perspective. We recognise that there remains much to do to better characterise the system and develop longer term co-culture systems where the survival of bacterial mutation can be explored. Mycobacteria can exit amoebae via a range of mechanisms including exocytosis, direct lysis, and non-lytic ejection [17, 87, 88]. Further work will characterise the mechanisms of amoeba exit used by wild-type M. bovis, and how the gene families we have identified influence each of these.
In summary, we have demonstrated that M. bovis is resistant to uptake and destruction by environmental D. discoideum and actively transits through the amoeba using a programme of mechanisms, many of which have been co-opted as virulence factors in the mammalian host. This, combined with the ability to replicate and adapt at ambient temperatures, may enhance the capacity of M. bovis to utilise an environmental stage between mammalian hosts and so enhance transmissibility. Understanding the transmission of M. bovis is crucial for implementing effective control measures, and understanding the survival of M. bovis outside of mammalian hosts warrants further investigation.
Data availability
The HiSeq datasets are available in the Sequence Read Archive (SRA) repository, Bioproject accession number PRJNA588141.
References
Allen AR, Skuce RA, Byrne AW. Bovine tuberculosis in Britain and Ireland—a perfect storm? the confluence of potential ecological and epidemiological impediments to controlling a chronic infectious disease. Front Vet Sci. 2018;5:109.
Brooks-Pollock E, Roberts GO, Keeling MJ. A dynamic model of bovine tuberculosis spread and control in Great Britain. Nature. 2014;511:228–31.
Olea-Popelka F, Muwonge A, Perera A, Dean AS, Mumford E, Erlacher-Vindel E, et al. Zoonotic tuberculosis in human beings caused by Mycobacterium bovis-a call for action. Lancet Infect Dis. 2017;17:e21–e5.
World Health Organization (WHO), Food and Agriculture Organization of the United Nations (FAO) and World Organisation for Animal Health (OIE). Roadmap for zoonotic tuberculosis. 2017.
Corner LA. The role of wild animal populations in the epidemiology of tuberculosis in domestic animals: how to assess the risk. Vet Microbiol. 2006;112:303–12.
Santos N, Almeida V, Gortazar C, Correia-Neves M. Patterns of Mycobacterium tuberculosis-complex excretion and characterization of super-shedders in naturally-infected wild boar and red deer. Vet Res. 2015;46:129.
Corner LA, Murphy D, Gormley E. Mycobacterium bovis infection in the Eurasian badger (Meles meles): the disease, pathogenesis, epidemiology and control. J Comp Pathol. 2011;144:1–24.
Woodroffe R, Donnelly CA, Ham C, Jackson SY, Moyes K, Chapman K, et al. Badgers prefer cattle pasture but avoid cattle: implications for bovine tuberculosis control. Ecol Lett. 2016;19:1201–8.
Courtenay O, Reilly LA, Sweeney FP, Hibberd V, Bryan S, Ul-Hassan A, et al. Is Mycobacterium bovis in the environment important for the persistence of bovine tuberculosis? Biol Lett. 2006;2:460–2.
Sweeney FP, Courtenay O, Ul-Hassan A, Hibberd V, Reilly LA, Wellington EM. Immunomagnetic recovery of Mycobacterium bovis from naturally infected environmental samples. Lett Appl Microbiol. 2006;43:364–9.
Ghodbane R, Mba Medie F, Lepidi H, Nappez C, Drancourt M. Long-term survival of tuberculosis complex mycobacteria in soil. Microbiology. 2014;160:496–501.
Ward AI, Judge J, Delahay RJ. Farm husbandry and badger behaviour: opportunities to manage badger to cattle transmission of Mycobacterium bovis? Prev Vet Med. 2010;93:2–10.
Sapriel G, Brosch R. Shared pathogenomic patterns characterize a new phylotype, revealing transition toward host-adaptation long before speciation of mycobacterium tuberculosis. Genome Biol Evol. 2019;11:2420–38.
Stinear TP, Seemann T, Harrison PF, Jenkin GA, Davies JK, Johnson PD, et al. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res. 2008;18:729–41.
Chen G, Zhuchenko O, Kuspa A. Immune-like phagocyte activity in the social amoeba. Science. 2007;317:678–81.
Drancourt M. Looking in amoebae as a source of mycobacteria. Micro Pathog. 2014;77:119–24.
Hagedorn M, Rohde KH, Russell DG, Soldati T. Infection by tubercular mycobacteria is spread by nonlytic ejection from their amoeba hosts. Science. 2009;323:1729–33.
Mba Medie F, Ben Salah I, Henrissat B, Raoult D, Drancourt M. Mycobacterium tuberculosis complex mycobacteria as amoeba-resistant organisms. PLoS One. 2011;6:e20499.
Cosson P, Soldati T. Eat, kill or die: when amoeba meets bacteria. Curr Opin Microbiol. 2008;11:271–6.
Gao LY, Harb OS, Abu Kwaik Y. Utilization of similar mechanisms by Legionella pneumophila to parasitize two evolutionarily distant host cells, mammalian macrophages and protozoa. Infect Immun. 1997;65:4738–46.
Greub G, Raoult D. Microorganisms resistant to free-living amoebae. Clin Microbiol Rev. 2004;17:413–33.
Molmeret M, Horn M, Wagner M, Santic M, Abu Kwaik Y. Amoebae as training grounds for intracellular bacterial pathogens. Appl Environ Microbiol. 2005;71:20–8.
Segal G, Shuman HA. Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect Immun. 1999;67:2117–24.
Shiloh MU, Champion PA. To catch a killer. What can mycobacterial models teach us about Mycobacterium tuberculosis pathogenesis? Curr Opin Microbiol. 2010;13:86–92.
Stamm LM, Brown EJ. Mycobacterium marinum: the generalization and specialization of a pathogenic mycobacterium. Microbes Infect. 2004;6:1418–28.
Tobin DM, Ramakrishnan L. Comparative pathogenesis of Mycobacterium marinum and Mycobacterium tuberculosis. Cell Microbiol. 2008;10:1027–39.
Cirillo JD, Falkow S, Tompkins LS, Bermudez LE. Interaction of Mycobacterium avium with environmental amoebae enhances virulence. Infect Immun. 1997;65:3759–67.
Danelishvili L, Wu M, Stang B, Harriff M, Cirillo SL, Cirillo JD, et al. Identification of Mycobacterium avium pathogenicity island important for macrophage and amoeba infection. Proc Natl Acad Sci USA. 2007;104:11038–43.
Kennedy GM, Morisaki JH, Champion PA. Conserved mechanisms of Mycobacterium marinum pathogenesis within the environmental amoeba Acanthamoeba castellanii. Appl Environ Microbiol. 2012;78:2049–52.
Volkman HE, Clay H, Beery D, Chang JC, Sherman DR, Ramakrishnan L. Tuberculous granuloma formation is enhanced by a mycobacterium virulence determinant. PLoS Biol 2004;2:e367.
Weerdenburg EM, Abdallah AM, Rangkuti F, Abd El Ghany M, Otto TD, Adroub SA, et al. Genome-wide transposon mutagenesis indicates that Mycobacterium marinum customizes its virulence mechanisms for survival and replication in different hosts. Infect Immun. 2015;83:1778–88.
Brock DA, Haselkorn TS, Garcia JR, Bashir U, Douglas TE, Galloway J, et al. Diversity of free-living environmental bacteria and their interactions with a bactivorous amoeba. Front Cell Infect Microbiol. 2018;8:411.
Garnier T, Eiglmeier K, Camus JC, Medina N, Mansoor H, Pryor M, et al. The complete genome sequence of Mycobacterium bovis. Proc Natl Acad Sci USA. 2003;100:7877–82.
Mendum TA, Chandran A, Williams K, Vordermeier HM, Villarreal-Ramos B, Wu H, et al. Transposon libraries identify novel Mycobacterium bovis BCG genes involved in the dynamic interactions required for BCG to persist during in vivo passage in cattle. BMC Genom. 2019;20:431.
Bardarov S, Bardarov S Jr, Pavelka MS Jr, Sambandamurthy V, Larsen M, Tufariello J, et al. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology. 2002;148:3007–17.
Butler RE, Cihlarova V, Stewart GR. Effective generation of reactive oxygen species in the mycobacterial phagosome requires K+ efflux from the bacterium. Cell Microbiol. 2010;12:1186–93.
Hagedorn M, Soldati T. Flotillin and RacH modulate the intracellular immunity of Dictyostelium to Mycobacterium marinum infection. Cell Microbiol. 2007;9:2716–33.
Long JE, DeJesus M, Ward D, Baker RE, Ioerger T, Sassetti CM. Identifying essential genes in Mycobacterium tuberculosis by global phenotypic profiling. Methods Mol Biol. 2015;1279:79–95.
Mendum TA, Wu H, Kierzek AM, Stewart GR. Lipid metabolism and Type VII secretion systems dominate the genome scale virulence profile of Mycobacterium tuberculosis in human dendritic cells. BMC Genom. 2015;16:372.
DeJesus MA, Ambadipudi C, Baker R, Sassetti C, Ioerger TR. TRANSIT–a software tool for Himar1 TnSeq analysis. PLoS Comput Biol. 2015;11:e1004401.
DeJesus MA, Gerrick ER, Xu W, Park SW, Long JE, Boutte CC, et al. Comprehensive essentiality analysis of the Mycobacterium tuberculosis genome via saturating transposon mutagenesis. MBio. 2017;8:pii: e02133–16.
Abdallah AM, Gey van Pittius NC, Champion PA, Cox J, Luirink J, Vandenbroucke-Grauls CM, et al. Type VII secretion–mycobacteria show the way. Nat Rev Microbiol. 2007;5:883–91.
Gordon SV, Brosch R, Billault A, Garnier T, Eiglmeier K, Cole ST. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol Microbiol. 1999;32:643–55.
Mahairas GG, Sabo PJ, Hickey MJ, Singh DC, Stover CK. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J Bacteriol. 1996;178:1274–82.
Pym AS, Brodin P, Majlessi L, Brosch R, Demangel C, Williams A, et al. Recombinant BCG exporting ESAT-6 confers enhanced protection against tuberculosis. Nat Med. 2003;9:533–9.
Lewis KN, Liao R, Guinn KM, Hickey MJ, Smith S, Behr MA, et al. Deletion of RD1 from Mycobacterium tuberculosis mimics bacille Calmette-Guerin attenuation. J Infect Dis. 2003;187:117–23.
Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci USA. 2003;100:12989–94.
Wards BJ, de Lisle GW, Collins DM. An esat6 knockout mutant of Mycobacterium bovis produced by homologous recombination will contribute to the development of a live tuberculosis vaccine. Tube Lung Dis. 2000;80:185–9.
Augenstreich J, Arbues A, Simeone R, Haanappel E, Wegener A, Sayes F, et al. ESX-1 and phthiocerol dimycocerosates of Mycobacterium tuberculosis act in concert to cause phagosomal rupture and host cell apoptosis. Cell Microbiol. 2017;19:e12726.
Conrad WH, Osman MM, Shanahan JK, Chu F, Takaki KK, Cameron J, et al. Mycobacterial ESX-1 secretion system mediates host cell lysis through bacterium contact-dependent gross membrane disruptions. Proc Natl Acad Sci USA. 2017;114:1371–6.
Abdallah AM, Bestebroer J, Savage ND, de Punder K, van Zon M, Wilson L, et al. Mycobacterial secretion systems ESX-1 and ESX-5 play distinct roles in host cell death and inflammasome activation. J Immunol. 2011;187:4744–53.
Simeone R, Bobard A, Lippmann J, Bitter W, Majlessi L, Brosch R, et al. Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathog. 2012;8:e1002507.
Simeone R, Sayes F, Song O, Groschel MI, Brodin P, Brosch R, et al. Cytosolic access of Mycobacterium tuberculosis: critical impact of phagosomal acidification control and demonstration of occurrence in vivo. PLoS Pathog. 2015;11:e1004650.
Smith J, Manoranjan J, Pan M, Bohsali A, Xu J, Liu J, et al. Evidence for pore formation in host cell membranes by ESX-1-secreted ESAT-6 and its role in Mycobacterium marinum escape from the vacuole. Infect Immun. 2008;76:5478–87.
van der Wel N, Hava D, Houben D, Fluitsma D, van Zon M, Pierson J, et al. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell. 2007;129:1287–98.
de Jonge MI, Pehau-Arnaudet G, Fretz MM, Romain F, Bottai D, Brodin P, et al. ESAT-6 from Mycobacterium tuberculosis dissociates from its putative chaperone CFP-10 under acidic conditions and exhibits membrane-lysing activity. J Bacteriol. 2007;189:6028–34.
Francis RJ, Butler RE, Stewart GR. Mycobacterium tuberculosis ESAT-6 is a leukocidin causing Ca2+ influx, necrosis and neutrophil extracellular trap formation. Cell Death Dis. 2014;5:e1474.
Gao LY, Guo S, McLaughlin B, Morisaki H, Engel JN, Brown EJ. A mycobacterial virulence gene cluster extending RD1 is required for cytolysis, bacterial spreading and ESAT-6 secretion. Mol Microbiol. 2004;53:1677–93.
Hsu T, Hingley-Wilson SM, Chen B, Chen M, Dai AZ, Morin PM, et al. The primary mechanism of attenuation of bacillus Calmette-Guerin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc Natl Acad Sci USA. 2003;100:12420–5.
Fortune SM, Jaeger A, Sarracino DA, Chase MR, Sassetti CM, Sherman DR, et al. Mutually dependent secretion of proteins required for mycobacterial virulence. Proc Natl Acad Sci USA. 2005;102:10676–81.
Hunt DM, Sweeney NP, Mori L, Whalan RH, Comas I, Norman L, et al. Long-range transcriptional control of an operon necessary for virulence-critical ESX-1 secretion in Mycobacterium tuberculosis. J Bacteriol. 2012;194:2307–20.
MacGurn JA, Raghavan S, Stanley SA, Cox JS. A non-RD1 gene cluster is required for Snm secretion in Mycobacterium tuberculosis. Mol Microbiol. 2005;57:1653–63.
López-Jiménez AT, Hagedorn M, Delincé MJ, McKinney J, Soldati T. The developmental cycle of Dictyostelium discoideum ensures curing of a mycobacterial infection at both cell-autonomous level and by collaborative exclusion. bioRxiv. 2019:586263. https://doi.org/10.1101/586263.
Stewart GR, Patel J, Robertson BD, Rae A, Young DB. Mycobacterial mutants with defective control of phagosomal acidification. PLoS Pathog. 2005;1:269–78.
Griffin JE, Gawronski JD, Dejesus MA, Ioerger TR, Akerley BJ, Sassetti CM. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 2011;7:e1002251.
Feltcher ME, Gunawardena HP, Zulauf KE, Malik S, Griffin JE, Sassetti CM, et al. Label-free quantitative proteomics reveals a role for the mycobacterium tuberculosis SecA2 pathway in exporting solute binding proteins and Mce transporters to the cell wall. Mol Cell Proteom. 2015;14:1501–16.
Camacho LR, Ensergueix D, Perez E, Gicquel B, Guilhot C. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol Microbiol. 1999;34:257–67.
Barczak AK, Avraham R, Singh S, Luo SS, Zhang WR, Bray MA, et al. Systematic, multiparametric analysis of Mycobacterium tuberculosis intracellular infection offers insight into coordinated virulence. PLoS Pathog. 2017;13:e1006363.
Quigley J, Hughitt VK, Velikovsky CA, Mariuzza RA, El-Sayed NM, Briken V. The cell wall lipid PDIM contributes to phagosomal escape and host cell exit of mycobacterium tuberculosis. MBio. 2017;8:pii: e00148–17.
Simeone R, Leger M, Constant P, Malaga W, Marrakchi H, Daffe M, et al. Delineation of the roles of FadD22, FadD26 and FadD29 in the biosynthesis of phthiocerol dimycocerosates and related compounds in Mycobacterium tuberculosis. FEBS J. 2010;277:2715–25.
Namouchi A, Karboul A, Fabre M, Gutierrez MC, Mardassi H. Evolution of smooth tubercle Bacilli PE and PE_PGRS genes: evidence for a prominent role of recombination and imprint of positive selection. PLoS ONE. 2013;8:e64718.
Gouzy A, Larrouy-Maumus G, Wu TD, Peixoto A, Levillain F, Lugo-Villarino G, et al. Mycobacterium tuberculosis nitrogen assimilation and host colonization require aspartate. Nat Chem Biol. 2013;9:674–6.
Sanchez-Hidalgo A, Obregon-Henao A, Wheat WH, Jackson M, Gonzalez-Juarrero M. Mycobacterium bovis hosted by free-living-amoebae permits their long-term persistence survival outside of host mammalian cells and remain capable of transmitting disease to mice. Environ Microbiol. 2017;19:4010–21.
Taylor SJ, Ahonen LJ, de Leij FA, Dale JW. Infection of Acanthamoeba castellanii with mycobacterium bovis and M. bovis BCG and survival of M. bovis within the amoebae. Appl Environ Microbiol. 2003;69:4316–9.
Brennan MJ. The enigmatic PE/PPE multigene family of mycobacteria and tuberculosis vaccination. Infect Immun. 2017;85:pii: e00969–16.
Gey van Pittius NC, Sampson SL, Lee H, Kim Y, van Helden PD, Warren RM. Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the ESAT-6 (esx) gene cluster regions. BMC Evol Biol. 2006;6:95.
Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–44.
Fishbein S, van Wyk N, Warren RM, Sampson SL. Phylogeny to function: PE/PPE protein evolution and impact on Mycobacterium tuberculosis pathogenicity. Mol Microbiol. 2015;96:901–16.
Ates LS. New insights into the mycobacterial PE and PPE proteins provide a framework for future research. Mol Microbiol. 2019;1–18.
Pandey AK, Sassetti CM. Mycobacterial persistence requires the utilization of host cholesterol. Proc Natl Acad Sci USA. 2008;105:4376–80.
Barisch C, Paschke P, Hagedorn M, Maniak M, Soldati T. Lipid droplet dynamics at early stages of Mycobacterium marinum infection in Dictyostelium. Cell Microbiol. 2015;17:1332–49.
Middlebrook G, Coleman CM, Schaefer WB. Sulfolipid from virulent tubercle bacilli. Proc Natl Acad Sci USA. 1959;45:1801–4.
Goren MB. Sulfolipid I of Mycobacterium tuberculosis, strain H37Rv. I. Purification and properties. Biochim Biophys Acta. 1970;210:116–26.
Gonzalo Asensio J, Maia C, Ferrer NL, Barilone N, Laval F, Soto CY, et al. The virulence-associated two-component PhoP-PhoR system controls the biosynthesis of polyketide-derived lipids in Mycobacterium tuberculosis. J Biol Chem. 2006;281:1313–6.
Molle V, Kremer L. Division and cell envelope regulation by Ser/Thr phosphorylation: Mycobacterium shows the way. Mol Microbiol. 2010;75:1064–77.
Molle V, Reynolds RC, Alderwick LJ, Besra GS, Cozzone AJ, Futterer K, et al. EmbR2, a structural homologue of EmbR, inhibits the Mycobacterium tuberculosis kinase/substrate pair PknH/EmbR. Biochem J. 2008;410:309–17.
Cardenal-Munoz E, Barisch C, Lefrancois LH, Lopez-Jimenez AT, Soldati T. When dicty met myco, a (Not So) romantic story about one amoeba and its intracellular pathogen. Front Cell Infect Microbiol. 2017;7:529.
Delince MJ, Bureau JB, Lopez-Jimenez AT, Cosson P, Soldati T, McKinney JD. A microfluidic cell-trapping device for single-cell tracking of host-microbe interactions. Lab Chip. 2016;16:3276–85.
Clemmensen HS, Knudsen NPH, Rasmussen EM, Winkler J, Rosenkrands I, Ahmad A, et al. An attenuated Mycobacterium tuberculosis clinical strain with a defect in ESX-1 secretion induces minimal host immune responses and pathology. Sci Rep. 2017;7:46666.
Feltcher ME, Sullivan JT, Braunstein M. Protein export systems of Mycobacterium tuberculosis: novel targets for drug development? Future Microbiol. 2010;5:1581–97.
Groschel MI, Sayes F, Simeone R, Majlessi L, Brosch R. ESX secretion systems: mycobacterial evolution to counter host immunity. Nat Rev Microbiol. 2016;14:677–91.
Lou Y, Rybniker J, Sala C, Cole ST. EspC forms a filamentous structure in the cell envelope of Mycobacterium tuberculosis and impacts ESX-1 secretion. Mol Microbiol. 2017;103:26–38.
Acknowledgements
With thanks to Sebastien Kicka and Cristina Bosmani for their advice on cultivating D. discoideum and performing flow cytometry assays. Thanks to Jana Baron and James Barber for assistance with cultivating D. discoideum. We thank the National Centre for the Replacement, Refinement and Reduction of Animals in Research (NC3Rs) for funding this work (NC/M002012/1 to GRS and TS). This work was funded by the National Centre for the Replacement, Refinement and Reduction of Animals in Research (NC3Rs, NC/M002012/1 to GRS and TS).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Butler, R.E., Smith, A.A., Mendum, T.A. et al. Mycobacterium bovis uses the ESX-1 Type VII secretion system to escape predation by the soil-dwelling amoeba Dictyostelium discoideum. ISME J 14, 919–930 (2020). https://doi.org/10.1038/s41396-019-0572-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-019-0572-z
This article is cited by
-
Predation-resistant Pseudomonas bacteria engage in symbiont-like behavior with the social amoeba Dictyostelium discoideum
The ISME Journal (2023)
-
The source and fate of Mycobacterium tuberculosis complex in wastewater and possible routes of transmission
BMC Public Health (2022)
-
Microbial warfare in the wild—the impact of protists on the evolution and virulence of bacterial pathogens
International Microbiology (2021)