Abstract
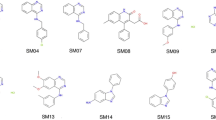
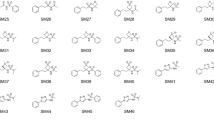
Partition coefficients describe the equilibrium partitioning of a single, defined charge state of a solute between two liquid phases in contact, typically a neutral solute. Octanol–water partition coefficients (\(K_{\rm ow}\)), or their logarithms (log P), are frequently used as a measure of lipophilicity in drug discovery. The partition coefficient is a physicochemical property that captures the thermodynamics of relative solvation between aqueous and nonpolar phases, and therefore provides an excellent test for physics-based computational models that predict properties of pharmaceutical relevance such as protein-ligand binding affinities or hydration/solvation free energies. The SAMPL6 Part II octanol–water partition coefficient prediction challenge used a subset of kinase inhibitor fragment-like compounds from the SAMPL6 \(\hbox {p}{K}_{{\rm a}}\) prediction challenge in a blind experimental benchmark. Following experimental data collection, the partition coefficient dataset was kept blinded until all predictions were collected from participating computational chemistry groups. A total of 91 submissions were received from 27 participating research groups. This paper presents the octanol–water log P dataset for this SAMPL6 Part II partition coefficient challenge, which consisted of 11 compounds (six 4-aminoquinazolines, two benzimidazole, one pyrazolo[3,4-d]pyrimidine, one pyridine, one 2-oxoquinoline substructure containing compounds) with log P values in the range of 1.95–4.09. We describe the potentiometric log P measurement protocol used to collect this dataset using a Sirius T3, discuss the limitations of this experimental approach, and share suggestions for future log P data collection efforts for the evaluation of computational methods.





Similar content being viewed by others
Data availability statement
All SAMPL6 log P Challenge instructions, submissions, experimental data and analysis are available at https://github.com/samplchallenges/SAMPL6/tree/master/physical_properties/logP
Notes
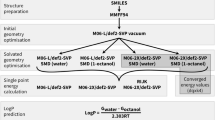
SAMPL6 was originally announced as featuring a log D prediction challenge, but there were difficulties in the collection of experimental data. The original plan was to measure log P0, log P−1, and log P+1 and calculate log D values at the experimental pH using these values. However, we were able to measure the partition coefficients of neutral species (log P0) much more reliably than ionic species with potentiometric log P method of Sirius T3, as elaborated further below.
Abbreviations
- SAMPL:
-
Statistical Assessment of the Modeling of Proteins and Ligands
- log P :
-
log\(_{10}\) of the organic solvent-water partition coefficient (\(K_{ow}\), refers to partition of neutral species unless stated otherwise)
- log D :
-
log\(_{10}\) of organic solvent-water distribution coefficient (\(D_{ow}\))
- log R :
-
log\(_{10}\) of the volumetric ratios of partition solvents (octanol to water)
- \(\hbox {p}{K}_{{\rm a}}\) :
-
−log\(_{10}\) of the acid dissociation equilibrium constant
- \(\hbox {p}_{{\rm o}}{K}_{{\rm a}}\) :
-
−log\(_{10}\) apparent acid dissociation equilibrium constant in octanol–water biphasic system
- ISA:
-
Ionic-strength adjusted solution with 0.15 M KCl
- SEM:
-
Standard error of the mean
- LC-MS:
-
Liquid chromatography-mass spectrometry
- NMR:
-
Nuclear magnetic resonance spectroscopy
- HRMS:
-
High-resolution mass spectrometry
- octanol:
-
1-Octanol, also known as n-octanol
References
Mobley DL, Chodera JD, Isaacs L, Gibb BC (2016) Advancing predictive modeling through focused development of model systems to drive new modeling innovations. UC Irvine: Department of Pharmaceutical Sciences, UCI. https://escholarship.org/uc/item/7cf8c6cr
Nicholls A, Mobley DL, Guthrie JP, Chodera JD, Bayly CI, Cooper MD, Pande VS (2008) Predicting small-molecule solvation free energies: an informal blind test for computational chemistry. J Med Chem 51(4):769–779. https://doi.org/10.1021/jm070549+
Guthrie JP (2009) A blind challenge for computational solvation free energies: introduction and overview. J Phys Chem B 113(14):4501–4507. https://doi.org/10.1021/jp806724u
Skillman AG, Geballe MT, Nicholls A (2010) SAMPL2 challenge: prediction of solvation energies and tautomer ratios. J Comput Aided Mol Des 24(4):257–258. https://doi.org/10.1007/s10822-010-9358-0
Geballe MT, Skillman AG, Nicholls A, Guthrie JP, Taylor PJ (2010) The SAMPL2 blind prediction challenge: introduction and overview. J Comput Aided Mol Des 24(4):259–279. https://doi.org/10.1007/s10822-010-9350-8
Skillman AG (2012) SAMPL3: blinded prediction of host–guest binding affinities, hydration free energies, and trypsin inhibitors. J Comput Aided Mol Des 26(5):473–474. https://doi.org/10.1007/s10822-012-9580-z
Geballe MT, Guthrie JP (2012) The SAMPL3 blind prediction challenge: transfer energy overview. J Comput Aided Mol Des 26(5):489–496. https://doi.org/10.1007/s10822-012-9568-8
Muddana HS, Varnado CD, Bielawski CW, Urbach AR, Isaacs L, Geballe MT, Gilson MK (2012) Blind prediction of host-guest binding affinities: a new SAMPL3 challenge. J Comput Aided Mol Des 26(5):475–487. https://doi.org/10.1007/s10822-012-9554-1
Guthrie JP (2014) SAMPL4, a blind challenge for computational solvation free energies: the compounds considered. J Comput Aided Mol Des 28(3):151–168. https://doi.org/10.1007/s10822-014-9738-y
Mobley DL, Wymer KL, Lim NM, Guthrie JP (2014) Blind prediction of solvation free energies from the SAMPL4 challenge. J Comput Aided Mol Des 28(3):135–150. https://doi.org/10.1007/s10822-014-9718-2
Muddana HS, Fenley AT, Mobley DL, Gilson MK (2014) The SAMPL4 host-guest blind prediction challenge: an overview. J Comput Aided Mol Des 28(4):305–317. https://doi.org/10.1007/s10822-014-9735-1
Mobley DL, Liu S, Lim NM, Wymer KL, Perryman AL, Forli S, Deng N, Su J, Branson K, Olson AJ (2014) Blind prediction of HIV integrase binding from the SAMPL4 challenge. J Comput Aided Mol Des 28(4):327–345. https://doi.org/10.1007/s10822-014-9723-5
Yin J, Henriksen NM, Slochower DR, Shirts MR, Chiu MW, Mobley DL, Gilson MK (2017) Overview of the SAMPL5 host-guest challenge: are we doing better? J Comput Aided Mol Des 31(1):1–19. https://doi.org/10.1007/s10822-016-9974-4
Bannan CC, Burley KH, Chiu M, Shirts MR, Gilson MK, Mobley DL (2016) Blind prediction of cyclohexane–water distribution coefficients from the SAMPL5 challenge. J Comput Aided Mol Des 30(11):1–18. https://doi.org/10.1007/s10822-016-9954-8
Rizzi A, Murkli S, McNeill JN, Yao W, Sullivan M, Gilson MK, Chiu MW, Isaacs L, Gibb BC, Mobley DL et al (2018) Overview of the SAMPL6 host-guest binding affinity prediction challenge. J Comput Aided Mol Des 32(10):937–963
Muddana HS, Gilson MK (2012) Prediction of SAMPL3 host-guest binding affinities: evaluating the accuracy of generalized force-fields. J Comput Aided Mol Des 26(5):517–525. https://doi.org/10.1007/s10822-012-9544-3
Gibb CLD, Gibb BC (2013) Binding of cyclic carboxylates to octa-acid deep-cavity cavitand. J Comput Aided Mol Des 28(4):319–325. https://doi.org/10.1007/s10822-013-9690-2
Cao L, Isaacs L (2014) Absolute and relative binding affinity of cucurbit[7]uril towards a series of cationic guests. Supramol Chem 26(3–4):251–258. https://doi.org/10.1080/10610278.2013.852674
Sullivan MR, Sokkalingam P, Nguyen T, Donahue JP, Gibb BC (2017) Binding of carboxylate and trimethylammonium salts to octa-acid and TEMOA deep-cavity cavitands. J Comput Aided Mol Des 31(1):1–8. https://doi.org/10.1007/s10822-016-9925-0
Bannan CC, Burley KH, Chiu M, Shirts MR, Gilson MK, Mobley DL (2016) Blind prediction of cyclohexane-water distribution coefficients from the SAMPL5 challenge. J Comput Aided Mol Des 30(11):927–944. https://doi.org/10.1007/s10822-016-9954-8
Rustenburg AS, Dancer J, Lin B, Feng JA, Ortwine DF, Mobley DL, Chodera JD (2016) Measuring experimental cyclohexane-water distribution coefficients for the SAMPL5 challenge. J Comput Aided Mol Des 30(11):945–958. https://doi.org/10.1007/s10822-016-9971-7
Waring MJ (2010) Lipophilicity in drug discovery. Expert Opin Drug Discov 5(3):235–248. https://doi.org/10.1517/17460441003605098
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (1997) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 23(1):3–25. https://doi.org/10.1016/S0169-409X(96)00423-1
Leo A, Hansch C, Elkins D (1971) Partition coefficients and their uses. Chem Rev 71(6):525–616. https://doi.org/10.1021/cr60274a001
Arnott JA, Planey SL (2012) The influence of lipophilicity in drug discovery and design. Expert Opin Drug Discov 7(10):863–875. https://doi.org/10.1517/17460441.2012.714363
Lipnick RL (2008) Environmental hazard assessment using lipophilicity data. In: Pliška V, Testa B, van de Waterbeemd H (eds) Methods and principles in medicinal chemistry. Wiley-VCH Verlag GmbH, Weinheim, pp 339–353. https://doi.org/10.1002/9783527614998.ch19
Pickard FC, König G, Tofoleanu F, Lee J, Simmonett AC, Shao Y, Ponder JW, Brooks BR (2016) Blind prediction of distribution in the SAMPL5 challenge with QM based protomer and pKa corrections. J Comput Aided Mol Des 30(11):1–14. https://doi.org/10.1007/s10822-016-9955-7
Işık M, Bergazin TD, Fox T, Rizzi A, Chodera JD, Mobley DL (2019) Assessing the accuracy of octanol-water partition coefficient predictions in the SAMPL6 Part II logP challenge. J Comput Aided Mol Des (SAMPL6 Part II special issue)
Işık M, Levorse D, Rustenburg AS, Ndukwe IE, Wang H, Wang X, Reibarkh M, Martin GE, Makarov AA, Mobley DL, Rhodes T, Chodera JD (2018) pKa measurements for the SAMPL6 prediction challenge for a set of kinase inhibitor-like fragments. J Comput Aided Mol Des 32(10):1117–1138. https://doi.org/10.1007/s10822-018-0168-0
Tsuji A, Kubo O, Miyamoto E, Yamana T (1977) Physicochemical properties of \(\beta\)-lactam antibiotics: oil-water distribution. J Pharm Sci 66(12):1675–1679. https://doi.org/10.1002/jps.2600661205
Avdeef A, Box KJ, Comer JEA, Hibbert C, Tam KY (1998) pH-Metric logP 10. Determination of liposomal membrane-water partition coefficients of lonizable drugs. Pharm Res 15(2):209–215. https://doi.org/10.1023/A:1011954332221
Octanol-Water Partitioning (2012) In: Absorption and drug development. Wiley, Hoboken pp 174–219. https://doi.org/10.1002/9781118286067.ch4
OECD (1995) Test No. 107: partition coefficient (n-octanol/water): shake flask method, OECD guidelines for the testing of chemicals, section 1. OECD Publishing, Paris. https://doi.org/10.1787/9789264069626-en
Hitzel L, Watt AP, Locker KL (2000) An increased throughput method for the determination of partition coefficients. Pharm Res 17(11):1389–1395. https://doi.org/10.1023/A:1007546905874
Lin B, Pease JH (2013) A novel method for high throughput lipophilicity determination by microscale shake flask and liquid chromatography tandem mass spectrometry. Comb Chem High Throughput Screen 16(10):817–825. https://doi.org/10.2174/1386207311301010007
De Bruijn J, Busser F, Seinen W, Hermens J (1989) Determination of octanol/water partition coefficients for hydrophobic organic chemicals with the “slow-stirring” method. Environ Toxicol Chem 8(6):499–512. https://doi.org/10.1002/etc.5620080607
Andersson JT, Schräder W (1999) A method for measuring 1-octanol water partition coefficients. Anal Chem 71(16):3610–3614. https://doi.org/10.1021/ac9902291
Herbert BJ, Dorsey JG (1995) n-Octanol-water partition coefficient estimation by micellar electrokinetic capillary chromatography. Anal Chem 67(4):744–749. https://doi.org/10.1021/ac00100a009
Berthod A, Carda-Broch S (2004) Determination of liquid-liquid partition coefficients by separation methods. J Chromatogr A 1037(1–2):3–14. https://doi.org/10.1016/j.chroma.2004.01.001
McCall JM (1975) Liquid-liquid partition coefficients by high-pressure liquid chromatography. J Med Chem 18(6):549–552. https://doi.org/10.1021/jm00240a003
Unger SH, Cook JR, Hollenberg JS (1978) Simple procedure for determining octanol aqueous partition, distribution, and ionization coefficients by reversed-phase high-pressure liquid chromatography. J Pharm Sci 67(10):1364–1367. https://doi.org/10.1002/jps.2600671008
Valkó K, Bevan C, Reynolds D (1997) Chromatographic hydrophobicity index by fast-gradient RP-HPLC: a high-throughput alternative to log P/log D. Anal Chem 69(11):2022–2029. https://doi.org/10.1021/ac961242d
Valkó K (1984) General approach for the estimation of octanol/water partition coefficient by reversed-phase high-performance liquid chromatography. J Liq Chromatogr 7(7):1405–1424. https://doi.org/10.1080/01483918408074054
OECD (2004) Test No. 117: partition coefficient (n-octanol/water): HPLC method, OECD guidelines for the testing of chemicals, section 1. OECD Publishing, Paris. https://doi.org/10.1787/9789264069824-en
Takacs-Novak K, Avdeef A (1996) Interlaboratory study of log P determination by shake-flask and potentiometric methods. J Pharm Biomed Anal 14:1405–1413
Avdeef A (1992) PH-metric log P. Part 1. Difference plots for determining ion-pair octanol-water partition coefficients of multiprotic substances. Quant Struct Act Relatsh 11(4):510–517
Sirius T3 User Manual, v1.1 (2008) Sirius Analytical Instruments Ltd. East Sussex, UK
Avdeef A (1993) pH-metric log P. II: refinement of partition coefficients and lonization constants of multiprotic substances. J Pharm Sci 82(2):183–190
Slater B, McCormack A, Avdeef A, Comer JE (1994) Ph-metric log P. 4. Comparison of partition coefficients determined by HPLC and potentiometric methods to literature values. J Pharm Sci 83(9):1280–1283
Comer J, Tam K (2001) Lipophilicity profiles: theory and measurement. Wiley-VCH, Zürich
OECD (2000) Guideline 122: partition coefficient (n-octanol/Water), pH-metric method for ionisable substances. Revised Draft
Işık M, Rustenburg A, Rizzi A, Bannan C, Gunner MR, Mobley DL, Chodera JD (2019) Accuracy of macroscopic and microscopic pKa predictions of small molecules evaluated by the SAMPL6 blind prediction challenge. J Comput Aided Mol Des. Manuscript in preparation
Wishart DS (2006) DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res 34(90001):D668–D672. https://doi.org/10.1093/nar/gkj067
Pence HE, Williams A (2010) ChemSpider: an online chemical information resource. J Chem Educ 87(11):1123–1124. https://doi.org/10.1021/ed100697w
NCI Open Database, August 2006 Release. https://cactus.nci.nih.gov/download/nci/
Enhanced NCI Database Browser 2.2. https://cactus.nci.nih.gov/ncidb2.2/
Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, Han L, He J, He S, Shoemaker BA, Wang J, Yu B, Zhang J, Bryant SH (2016) PubChem substance and compound databases. Nucleic Acids Res 44(D1):D1202–D1213. https://doi.org/10.1093/nar/gkv951
Tam KY, Takács-Novák K (2001 Apr) Multi-wavelength spectrophotometric determination of acid dissociation constants: a validation study. Anal Chim Acta 434(1):157–167. https://doi.org/10.1016/S0003-2670(01)00810-8
Allen RI, Box KJ, Comer JEA, Peake C, Tam KY (1998) Multiwavelength spectrophotometric determination of acid dissociation constants of ionizable drugs. J Pharm Biomed Anal 17(4–5):699–712. https://doi.org/10.1016/S0731-7085(98)00010-7
Ahn S, Fessler JA (2003) Standard errors of mean, variance, and standard deviation estimators. EECS Department, The University of Michigan. pp 1–2. http://web.eecs.umich.edu/~fessler/papers/lists/files/tr/stderr.pdf
OEDepict Toolkit 2017. Feb. 1. OpenEye Scientific Software, Santa Fe, NM. http://www.eyesopen.com
Takács-Novák K, Avdeef A, Box KJ, Podányi B, Szász G (1994) Determination of protonation macro- and microconstants and octanol/water partition coefficient of the antiinflammatory drug niflumic acid. J Pharm Biomed Anal 12(11):1369–1377. https://doi.org/10.1016/0731-7085(94)00090-5
Port A, Bordas M, Enrech R, Pascual R, Rosés M, Ràfols C, Subirats X, Bosch E (2018) Critical comparison of shake-flask, potentiometric and chromatographic methods for lipophilicity evaluation (Log P o/w ) of neutral, acidic, basic, amphoteric, and zwitterionic drugs. Eur J Pharm Sci 122:331–340. https://doi.org/10.1016/j.ejps.2018.07.010
Tielker N, Tomazic D, Eberlein L, Güssregen S, Kast SM (2019) The SAMPL6 challenge on predicting octanol-water partition coefficients from ECRISM theory. J Comput Aided Mol Des (SAMPL6 Part II special issue)
Acknowledgements
MI and JDC acknowledge support from the Sloan Kettering Institute. JDC acknowledges partial support from NIH Grant P30 CA008748. MI, JDC, ASR, and DLM gratefully acknowledge support from NIH Grant R01GM124270 supporting the SAMPL Blind Challenges. MI acknowledges support from a Doris J. Hutchinson Fellowship. DLM appreciates financial support from the National Institutes of Health (1R01GM108889-01), the National Science Foundation (CHE 1352608). The authors are extremely grateful for the assistance and support from the MRL Preformulations and NMR Structure Elucidation groups for materials, expertise, and instrument time, without which this SAMPL challenge would not have been possible. The authors would like to thank Ryan Cohen from the NMR Structure Elucidation group for the NMR and LC-MS analysis of SM13. MI and DL are grateful to Pion/Sirius Analytical for their technical support in the planning and execution of this study. We are especially thankful to Karl Box (Sirius Analytical) for the guidance on optimization and interpretation of log P measurements with the Sirius T3. We thank Brad Sherborne (MRL; ORCID: 0000-0002-0037-3427) for his valuable insights at the conception of the log P challenge and connecting us with TR and DL who were able to provide resources for experimental measurements. We acknowledge contributions from Caitlin Bannan who provided feedback on experimental data collection and structure of log P challenge from a computational chemist’s perspective. MI and JDC are grateful to OpenEye Scientific for providing a free academic software license for use in this work. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank anonymous reviewers for their input and constructive comments that improved this manuscript. Research reported in this publication was supported by National Institute for General Medical Sciences of the National Institutes of Health under Award Number R01GM124270 and R01GM108889, and from the National Cancer Institute of the National Institutes of Health under P30CA008748.
Disclaimers
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
Conceptualization, MI, JDC, TR, DLM ; Methodology, MI, DL; Software, MI ; Formal Analysis, MI ; Investigation, MI, DL; Resources, TR, DL; Data Curation, MI ; Writing-Original Draft, MI, DL; Writing - Review and Editing, MI, DL, JDC, TR, DLM; Visualization, MI ; Supervision, JDC, TR, DLM ; Project Administration, MI ; Funding Acquisition, DLM, JDC, TR, MI.
Corresponding authors
Ethics declarations
Conflict of interest
JDC was a member of the Scientific Advisory Board for Schrödinger, LLC during part of this study. JDC and DLM are current members of the Scientific Advisory Board of OpenEye Scientific Software. The Chodera laboratory receives or has received funding from multiple sources, including the National Institutes of Health, the National Science Foundation, the Parker Institute for Cancer Immunotherapy, Relay Therapeutics, Entasis Therapeutics, Silicon Therapeutics, EMD Serono (Merck KGaA), AstraZeneca, Vir Biosciences, XtalPi, the Molecular Sciences Software Institute, the Starr Cancer Consortium, the Open Force Field Consortium, Cycle for Survival, a Louis V. Gerstner Young Investigator Award, The Einstein Foundation, and the Sloan Kettering Institute. A complete list of funding can be found at http://choderalab.org/funding.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Işık, M., Levorse, D., Mobley, D.L. et al. Octanol–water partition coefficient measurements for the SAMPL6 blind prediction challenge. J Comput Aided Mol Des 34, 405–420 (2020). https://doi.org/10.1007/s10822-019-00271-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-019-00271-3