Abstract
Current prevention methods for the transmission of Mycobacterium leprae, the causative agent of leprosy, are inadequate as suggested by the rate of new leprosy cases reported. Simple large-scale detection methods for M. leprae infection are crucial for early detection of leprosy and disease control. The present study investigates the production and seroreactivity of a recombinant polypeptide composed of various M. leprae protein epitopes. The structural and physicochemical parameters of this construction were assessed using in silico tools. Parameters like subcellular localization, presence of signal peptide, primary, secondary, and tertiary structures, and 3D model were ascertained using several bioinformatics tools. The resultant purified recombinant polypeptide, designated rMLP15, is composed of 15 peptides from six selected M. leprae proteins (ML1358, ML2055, ML0885, ML1811, ML1812, and ML1214) that induce T cell reactivity in leprosy patients from different hyperendemic regions. Using rMLP15 as the antigen, sera from 24 positive patients and 14 healthy controls were evaluated for reactivity via ELISA. ELISA-rMLP15 was able to diagnose 79.17% of leprosy patients with a specificity of 92.86%. rMLP15 was also able to detect the multibacillary and paucibacillary patients in the same proportions, a desirable addition in the leprosy diagnosis. These results summarily indicate the utility of the recombinant protein rMLP15 in the diagnosis of leprosy and the future development of a viable screening test.
Similar content being viewed by others
Introduction
The chronic infectious disease leprosy is caused by the intracellular, acid-fast bacillus known as Mycobacterium leprae (Araújo 2003). M. leprae may cause dermatological and neurological granulomatous lesions on the skin that may lead to varying levels of numbness and incapacitation (Porto et al. 2015). Despite declining numbers of global leprosy cases, the disease is still endemic to many countries, with Brazil, in particular, ranking the second highest in the number of new cases reported (22,940 in 2017 alone) (Vieira et al. 2018). The World Health Organization (WHO) has delineated objectives to stop the transmission of new leprosy cases between 2016 and 2020. Among them, the development of new diagnostic tools is emphasized to be of utmost importance (WHO 2016). Additionally, the WHO proposes a standardized screening and treatment protocol by introducing an operational classification of multibacillary (MB) leprosy upon a positive smear test, regardless of the number of lesions (Reibel et al. 2015).
Well-trained clinicians able to identify clinical signs and symptoms in patients are crucial for an accurate diagnosis of leprosy (Richardus et al. 2017). Delayed diagnosis occurs frequently though, owing to few available clinical experts in the field (Corstjens et al. 2019), and increases the risk of severe disabilities (van Hooij et al. 2019). Other diagnostic methods like bacilloscopy and histopathology also lack adequate sensitivity and rely on well-trained technicians as well (Cheng et al. 2019). Molecular diagnostic methods like PCR and qPCR are difficult and expensive to perform in the field, despite having high levels of sensitivity (Martinez et al. 2014; Cheng et al. 2019). Although serological tests based on M. leprae antigens are available, they lack adequate sensitivity and are only for supporting clinical diagnosis (Kim et al. 2013). Although primarily used for detecting MB patients, the phenolic glycolipid I (PGL-I) (Roche et al. 1999) and the Leprosy IDRI Diagnostic-1 (LID-I) tests stand out (Duthie et al. 2007; Hungria et al. 2012). Also of significance is the NDO-LID® test, a rapid serological, lateral flow test designed with two proteins, ND-O (a synthetic PGL-I mimetic) and LID-I (a fusion protein of ML0405 and ML2331) (Reece et al. 2006; Hungria et al. 2017; van Hooij et al. 2018).
A number of M. leprae proteins and subsequently, tests based on these proteins, have been developed since elucidation of its genomic sequence (Cole et al. 2001) for serological diagnosis of leprosy (Meeker et al. 1986; Duthie et al. 2007; Hungria et al. 2017). These tests could only detect lepromatous and symptomatic cases, but not paucibacillary (PB) cases (Kumar et al. 2014; Duthie et al. 2014; Bahmanyar et al. 2016). The spectrum of outcomes following M. leprae infection is determined by host factors (van Hooij et al. 2019) ranging from anti-inflammatory T helper-2 (Th2)-mediated immunity against high bacterial loads and antibodies against M. leprae antigens in MB leprosy to strong pro-inflammatory T helper-1 (Th1) and T helper-17 (Th17)-mediated immunity characteristic of PB leprosy (Saini et al. 2013). The human leukocyte antigen (HLA) alleles are also hypothesized to influence host immune responses against M. leprae infection (de Souza-Santana et al. 2015). Thus, a reliable diagnostic test for leprosy should be able to capture the different clinical outcomes of M. leprae infection, including both cellular and humoral markers (van Hooij et al. 2019).
In a study by Bobosha et al. (2012), epitopes were identified and synthesized from a virulent group of M. leprae proteins with predicted promiscuous binding affinities to HLA class I or II alleles. Immunogenicity was tested using peripheral blood mononuclear cells (PBMCs) or whole blood isolated from patients and healthy endemic controls (HCs) from Brazil, Ethiopia, and Nepal. T-cell reactivity was induced in some hyperendemic patients without inducing cross-reactivity with other Mycobacterium species. In light of these results, we propose that unique candidate peptides of M. leprae could act as more precise diagnostic targets to measure, alongside the cellular and humoral immune responses. Our hypothesis that the inclusion of epitopes from high T-cell reactive proteins of M. leprae to the protein might lead to a better antibody response due to T-cell dependent B-cell activation.
Thus, the current study aimed to generate a single recombinant polypeptide composed of epitopes from high T-cell reactive proteins of M. leprae (Bobosha et al. 2012) and validate its seroreactivity in leprosy patients. This is based on previous reports to produce a synthetic protein that combines highly reactive segments of M. leprae antigens within a single product.
Materials and methods
DNA sequence construction of recombinant polypeptide MLP15
High T-cell reactive epitopes of 15 peptides from six different M. leprae proteins studied previously (Bobosha et al. 2012) were combined into a recombinant polypeptide designated rMLP15. Multiple epitopes showing HLA I/II reactivity were also placed at random intervals in the polypeptide. Selected epitopes were separated by 3-glycine linker residues and the Vector-NTI Express v1.1.1 program was used for designing a codon-optimized DNA sequence (for Escherichia coli, Epoch Life Science, Houston, TX, USA) of the polypeptide rMLP15 (GenBank accession number MN178257). NCBI’s Blastp (Protein BLAST, Altschul et al. 1990) was then used to analyze and exclude cross-reactive protein fragments of genus Mycobacterium (taxid: 81858) and Homo sapiens (taxid: 9606).
In silico analyses
The predictions of physicochemical properties were performed using ProtParam tool (Gasteiger et al. 2005) from ExPASy, which provided molecular weight, theoretical isoelectric point (pI), amino acid composition, extinction coefficient, estimated half-life, instability, and aliphatic indexes. The RaptorX software and web server were used for the estimation of primary, secondary, tertiary structures, and 3D models of the polypeptide (Källberg et al. 2012) and edited using Discovery Studio Visualization -BIOVIA (2017).
Subcellular localization of the sequence was assessed using three online software packages: PSORTb 3.0.2 (Yu et al. 2010), CELLO v.2.5 (Yu et al. 2004), and Gneg-mPLoc (Shen and Chou 2010). Consistent results in two out of three software predictions were considered. Signal peptides for cleavage were checked for using SignalP 4.1 (Petersen et al. 2011), and VaxiJen v2.0 was used for the prediction of protective antigens (Doytchinova and Flower 2007).
Cloning, expression, and purification of polypeptide MLP15
Recombinant DNA sequences encoding rMLP15 were cloned in pAE expression vector (Ramos et al. 2004) using the restriction enzyme sites BamHI and HindIII. The resulting plasmid pAE/rMLP15 was used to transform E. coli BL21 Star™ (DE3) strain (Invitrogen, Carlsbad, CA, USA). Following verification of expression with a small-scale test, one colony was inoculated with 500 mL of lysogeny broth (LB, Bertani 1951). rMLP15 expression was induced by adding 1 M isopropyl β-d-1-thiogalactopyranoside (IPTG) to the culture (Sigma-Aldrich, St. Louis, MO, USA) for 3 h at 37 °C with agitation. Purification protocol was initiated with a brief sonication followed by centrifugation, in accordance with Simionatto et al. (2010). The supernatant containing the recombinant protein was solubilized in wash buffer (20 mM Tris–HCL, 8 M urea, 500 mM NaCl, 300 mM imidazole) at pH 8.0 (Simionatto et al. 2010). Affinity chromatography using HisTrap™ (GE Healthcare, Madison, WI, USA) 1 mL columns precharged with Ni-Sepharose was used to purify rMLP15. The concentration and purity of the purified rMLP15 were determined by the BCA Assay (Pierce, Rockford, IL, USA) and 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE), respectively.
Western blot with recombinant proteins
Western blot was performed with the purified rMLP15. After solubilizing in a sample buffer (62.5 mM Tris–HCl, 10% glycerol, 5% 2-β-mercaptoethanol, 2% SDS) at pH 6.8 and separated by 12% SDS–PAGE, proteins electroblotted onto nitrocellulose membranes (GE Healthcare, Madison, WI, USA) and incubated in 5% skim milk diluted in PBS-T (phosphate-buffered saline with Tween-20) overnight at 4 °C. The membranes were then washed thrice with PBS-T, and incubated for 1 h at 37 °C with an anti-6 × His-tag monoclonal antibody (Sigma-Aldrich, St. Louis, MO, USA) at a 1:6000 dilution in PBS-T. Following three PBS-T washes, the membranes were incubated with horseradish peroxidase (HRP)-conjugated mouse anti-IgG (Sigma-Aldrich, St. Louis, MO, USA), at 1:6000 and incubated for 1 h at 37 °C. DAB (3,3′-diaminobenzidine) was used for the visualization of reactive bands.
Study population
Of the 24 patients with leprosy from whom sera were obtained for evaluation with ELISA, 10 were multibacillary (MB) and 14 paucibacillary (PB). The patients with positive bacilloscopy were classified as MB irrespective of the number of lesions (Reibel et al. 2015). Patients were clinically diagnosed by a dermatologist at a referral center for leprosy diagnosis and classified by bacilloscopy through a smear. The sera from 14 healthy individuals, was used as control. They were from a low endemic region with negative clinical diagnosis of leprosy and with no contact with leprosy and tuberculosis patients. All participating in the study were older than 18 years. The blood samples were collected by venipuncture and sera, once separated, were stored at − 20 °C until use. All participants in this study signed informed consent forms prior to enrolment and the study was approved by the Research Ethics Committee (1.816.093–11/09/2016) of the Federal University of Grande Dourados (UFGD).
Enzyme-linked immunosorbent assay (ELISA)
The immunoreactivity of anti-rMLP15 IgG in patient sera was assessed using a modified ELISA protocol of Lima et al. (2017). In polystyrene 96-well ELISA plates (Kasvi™), 1 µg/well of rMLP15 in 100 μL of 0.1 M sodium carbonate buffer (pH 9.6) was coated and incubated for 16–18 h at 4 °C. Blocking was done with 200 µL/well of 5% skim milk in PBS-T for 2 h at 37 °C after washing the plates thrice with 0.05% PBS-T. Thereafter, the plates were washed three times with PBS-T and 100 µL/well of patient sera (diluted 1:50 in 5% skim milk in 0.05% PBS-T) was added. The plates were incubated at 37 °C and after three PBS-T washes, 100 μL/well of anti-human IgG peroxidase-conjugated antibody (Sigma-Aldrich, St. Louis, MO, USA) was added, diluted 1:10,000 in PBS-T, and further incubated at 37 °C for 1 h. The chromogenic reaction was developed by the addition of 100 μL/well of tetramethylbenzidine solution (Sigma-Aldrich, St. Louis, MO, USA) for 15 min at 37 °C, and the reaction was stopped with 100 μL/well of 2 N sulfuric acid. Absorbance at 450 nm was measured with a spectrophotometer (Bio-Rad, Hercules, CA, USA) and mean OD values were calculated from the serum samples assayed in triplicate.
Statistical analysis
Statistical analyses included one-way analysis of variance (ANOVA), with Dunn’s test for multiple comparisons, and assessed using GraphPad Prism v.5.0 software (GraphPad Inc., La Jolla, CA, USA). The parameters of sensitivity, specificity, accuracy, and cutoff value based on Youden’s index were determined using a receiver operating characteristic (ROC) curve. Statistical significance was set at p < 0.05.
Results
Construction of polypeptide rMLP15 and in silico analysis
The epitopes selected from six M. leprae virulent proteins for rMLP15 construction are presented in Table 1.
A 569 bp DNA sequence for the proposed rMLP15 was designed and translated into a 189 amino-acid polypeptide with a molecular weight of 17.3 kDa (Table 2). A BLASTp search for homologous proteins with Mycobacterium genera did not yield any results, indicating minimal cross-reactivity with other pathogens of the same genus. rMLP15 was not predicted to have premature cleavage or inadequate expression based on SignalP analysis, while predictions by the software packages yielded three possible intracellular localizations possible for rMLP15 (Table 2). The analysis by VaxiJen showed the possible use of rMLP15 in vaccine trials due to positivity for protective antigen (Table 2). A high pI of 10.5 and grand average of hydropathy (GRAVY index) score of 0.70 were found for rMLP15 using the ProtParam tool (Table 2).
Physicochemical parameters calculated by ProtParam, subcellular localization, signal peptide prediction and protective antigen prediction of MLP15. The software used is given in parentheses.
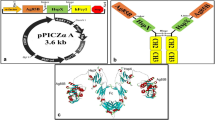
The results of primary, secondary, and tertiary structure analyses performed using RaptorX are presented in Fig. 1. The protein structure appears to contain two domains of amino acids 1–130 (domain 1) and 131–189 (domain 2) (Fig. 1a and b). The protein showed high accessibility of residues with 132 exposed amino acids and only 8 buried, besides 49 amino acids with medium exposure (Fig. 1c). rMLP15 shows a primary coil and turn conformation, and achieves structural extension and linearization owing to the glycine linkers and the majority of exposed amino acids (Fig. 1a–c). The analysis also reveals rMLP15 to be essentially hydrophobic, with 89 hydrophobic, 76 neutral, and 24 hydrophilic amino acid residues (Fig. 1d).
Primary structure and 3D rendering of MLP15. 3D rendering of MLP15. a The secondary structure of MLP15 indicating the location of the domains and turns and helices. b Tertiary extrusion prediction in the 3D model presenting the two main domains of MLP15. Blue: epitopes; and yellow: 3-Gly Linker. c Exposed surface of MLP15. Yellow: exposed amino acids; and blue: buried amino acids. d Hydrophobic surface of polypeptide MLP15 with color representation. Brown: more hydrophobic; blue: more hydrophilic; white: neutral. The analyses were done by RaptorX and edited in Discovery Studio Visualization-Biovia
Cloning, expression, and purification
The DNA sequence of MLP15 was cloned in a pAE expression vector and confirmed by restriction enzyme digestion, PCR, and sequencing. The highest level of MLP15 expression was obtained with the E. coli BL21 STAR (DE3) strain (Fig. 2). The protein yield obtained after purification was approximately 2.2 mg/L. After the purification, the presence of homodimers with approximately 35 kDa and monomers of 17 kDa (the predictable size of the rMPL15) were observed in western blot, using the anti-6 His tag (Fig. 2).
ELISA-rMLP15
The ELISA assays known for their proven diagnostic properties in leprosy were used to evaluate the performance of the recombinant antigen rMLP15. A ROC curve was constructed for the 24 patients and 14 HCs (Fig. 3), the cutoff value was based on the highest likelihood ratio (11.08) and the Youden index J was calculated (0.7203). rMLP15 showed an antibody response in 19 of 24 patients (79.17%) and 1 of 14 HCs (7.14%) from the sera evaluated in ELISA (Fig. 3a), thereby yielding a specificity rate of 92.86% (95% CI 66.13% to 99.82%), a sensitivity rate of 79.17% (95% CI 57.85% to 92.87%), and an area under the ROC curve (AUC) of 0.83 (95% CI 0.7006 to 0.9720) (Fig. 3b). In patients with leprosy, IgG levels were significantly elevated than in the control group (p < 0.001). Among patients with leprosy, rMLP15 was able to accurately identify 7 of 10 patients (70%) with MB and 12 of 14 PB patients (85.7%) (Fig. 3c) with a false positive rate of 7.14% (1 of 14 HCs). Statistical analysis did not reveal significant differences between the MB and PB groups, but both were significantly (p < 0.001) different from the HC group (Fig. 3d).
ELISA test performed with the sera from patients positive for leprosy using rMLP15. The results from indirect ELISA-rMLP15 for patient sera. a ELISA-rMLP15 in the sera from 24 leprosy patients and 14 healthy controls (HC). b Receiver operating characteristic (ROC) curve used to distinguish between positive and healthy controls (HC). c The levels of anti-rMLP15 IgG stratified paucibacillary patients (PB) and multibacillary patients (MB). d The levels of IgG anti-rMLP15 compared in HC (14), MB (10), and PB (14) patients; the serum levels were analyzed by ANOVA, followed by post hoc using Dunn’s test. **p < 0.001. AUC area under the curve, CI confidence interval, Se sensitivity, Sp specificity, LR likelihood ratio, PB paucibacillary, and MB multibacillary
ROC analysis was also performed on the subgroups of MB and PB patients (Fig. 4) and revealed a small increase in the cutoff values for MB patients, from 0.1993, in the overall analysis, to 0.2036 (Fig. 4a), while there was no change for PB data (Fig. 4c). Specificity was also unchanged for MB and PB subgroups in this analysis (92.86%). PB data showed a higher AUC value of 0.8827 (95% CI 0.7339 to 1.031) (Fig. 4d) than MB, 0.7714 (95% CI 0.5305 to 1.012) (Fig. 4b). The Youden index J was 0.6286 for MB and 0.7857 for PB.
ELISA test stratifying MB and PB patients using rMLP15. The results of indirect ELISA-rMLP15 for MB and PB patient sera. a ELISA-rMLP15 in sera from 10 leprosy MB patients and 14 HC. b Receiver operating characteristic (ROC) curve used to distinguish between positive MB and HC. c ELISA-MLP15 in sera from 14 leprosy PB patients and 14 HC. d Receiver operating characteristic (ROC) curve used to distinguish between positive PB and HC. AUC area under the curve, CI confidence interval, Se sensitivity, Sp specificity, LR likelihood ratio, HC healthy control, PB paucibacillary, and MB multibacillary
Discussion
The transmission of leprosy may be reduced by the use of tools that can accurately detect the disease at an early stage. Genomic sequencing has improved outcomes in this context by providing a more rational approach in the search of diagnostic tools for the diagnosis of many infectious diseases (Nagai et al. 2009). It has been established that the combined detection of humoral and cellular markers is efficient in diagnosing MB and PB leprosy patients (van Hooij et al. 2017, 2018). This study proposes a new design approach and a novel recombinant polypeptide for the serological detection of leprosy. A recombinant fusion protein based on six virulent M. leprae proteins (designated rMLP15), with established T-cell reactivity in leprosy patients, was used in an indirect ELISA to evaluate humoral responses in sera from leprosy-positive patients and healthy controls. Among the proteins that had epitopes selected for the construction of MLP15, ML2055 (5 epitopes) and ML1358 (5 epitopes), constituting 10/15 of MLP15, have been described as proteins with T cell and B cell epitopes that are immune reactive in the context of disease (Sampaio et al. 2011; Deval et al. 2016). rMLP15 demonstrated IgG reactivity in human sera for both MB and PB leprosy patients. These data confirm a plausible methodology that may be used to develop and improve new diagnostic tools for leprosy.
A number of bioinformatics analyses were carried out during the development process for rMLP15 to effectively guide the process and rationalize parameters to aid in better expression and purification of rMLP15. rMLP15 has been designed with some characteristics to optimize its use in diagnostics, like an extended chain structure with exposed amino acid residues to increase the possibility of recognition by specific immunoglobulins (Bergamaschi et al. 2019). The favored conformation of rMLP15 tertiary structure may be due to the ease of epitope–epitope interactions achieved by the presence of the 3-glycine linkers separating linear epitopes. Despite these optimizations, dimerization of the expressed rMLP15 was observed (Fig. 2), which may be due to intermolecular disulfide bond formation within homodimers or heterodimers and may result in higher oligomer formation (Futami et al. 2016). This could be explained by the high pI value (10.5) of rMLP15. It should be noted that this conformational observation does not seem to be a limiting factor in reactive antibody detection in leprosy patients.
The clinical course of disease progression in leprosy depends on the individual immune system (Alves et al. 2019). Genetic predisposition has been implicated to play a role in both disease susceptibility and host immune responses (Shankarkumar et al. 2003). The use of high-affinity HLA-binding linear epitopes in the construction of rMLP15 may aid in detecting both MB and PB leprosy patients. Available commercial tests like PGL-I or LID-1 protein-based tests can detect MB patients with high bacillary loads, but fail to detect PB leprosy patients, delegating them to a supportive test for treatment direction but not useful to detect early stages of the disease (Spencer et al. 2011; Geluk et al. 2011; van Hooij et al. 2017; Duthie et al. 2007; Leturiondo et al. 2019). PB leprosy, unlike MB leprosy, displays a dominant cellular phenotype with restricted anti-M. leprae antibody production. Our results show that rMLP15 was able to demonstrate seroreactivity in both PB and MB patients without significant differences in the levels of IgG antibodies between them.
rMLP15 demonstrated a higher detection rate of 79.17% (19 of 24) than by the NDO-LID® test (62.8%, Orange Life, Rio de Janeiro, Brazil) (Frade et al. 2017). The detection rates among available commercial tests vary widely depending on patient populations, as well as disease spectrum, with lower rates in PB leprosy patients (van Hooij et al. 2017). The detection rates found in a study that used proteins like NDO-LID-1 and PGL-1 were 34% and 32% in PB patients versus 73.6% and 81% in MB individuals, respectively (Leturiondo et al. 2019). Although this test was able to exhibit better detection capabilities than standard tests using PGL-I and LID-I, monitoring of individuals in the early stages of the disease and/or PB patients remains a challenge (Frade et al. 2017).
This is the first study, to the best of our knowledge, describing enhanced serological detection rates for PB patient cases. rMLP15 detected 85.71% (12 of 14) PB patients and 70% (7 of 10) of MB patients, both with a specificity of 92.86% (Fig. 4). Additionally, the false positive rate with rMLP15 was 7.14% (1 of 14), which is lower than that reported in the PGL-I based tests (> 10%) (Alban et al. 2014). Although reported from a limited number of samples, these results encourage us to propose this peptide as a potential tool for early detection of leprosy, especially in cases with undetectable bacterial loads and few clinical signs, like in the PB patients and household contacts, that have a 4 to 9 times greater risk of developing leprosy than the general population (van Beers et al. 1999). Early detection is important not only to facilitate diagnosis and classification but also for decision-making regarding the candidates for prophylactic interventions, a key strategy for disrupting the transmission of disease (van Hooij et al. 2019).
The detection approach followed in this study shows that rMLP15 is able to diagnose cases of leprosy with high sensitivity and specificity multibacillary and paucibacillary patients. Our results put forward rMLP15 as a potential tool for leprosy diagnosis and the availability of such recombinant polypeptides could simplify future diagnostic test development. Further studies are needed to better characterize rMLP15 including testing with patient samples from different endemic regions, as well as in sera collected from individuals coming in contact with these patients and evaluate their ability to induce cellular responses.
Availability of data and material
Not applicable
References
Alban SM, de Moura JF, Thomaz-Soccol V, Sékula SB, Alvarenga LM, Mira MT, Olortegui CC, Minozzo JC (2014) Phage display and synthetic peptides as promising biotechnological tools for the serological diagnosis of leprosy. PLoS ONE. https://doi.org/10.1371/journal.pone.0106222
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1016/s0022-2836(05)80360-2
Alves HV, de Moraes AG, Pepineli AC, Tiyo BT, de Lima Neto QA, da Silva Santos T, Teixeira JJV, Ambrosio-Albuquerque EP, Sell AM, Visentainer JEL (2019) The impact of KIR/HLA genes on the risk of developing multibacillary leprosy. PLoS Negl Trop Dis. https://doi.org/10.1371/journal.pntd.0007696
Araújo MG (2003) Leprosy in Brazil. Rev Soc Bras Med Trop 36:373–82 http://www.scielo.br/scielo.php?pid=S0037-6822003000300010&script=sci_arttext&tlng=pt
Bahmanyar ER, Smith WC, Brennan P, Cummings R, Duthie M, Richardus JH, Saunderson P, Shwe T, Rosen S, Geluk A (2016) Leprosy diagnostic test development as a prerequisite towards elimination: requirements from the user’s perspective. PLoS Negl Trop Dis 10:e0004331
Bergamaschi G, Fassi E, Romanato A, D’Annessa I, Odinolfi MT, Brambilla D, Damin F, Chiari M, Gori A, Colombo G (2019) Computational analysis of dengue virus envelope protein (E) reveals an epitope with Flavivirus immunodiagnostic potential in peptide microarrays. Int J Mol Sci 20:1921
Bobosha K, Tang ST, van der Ploeg-van Schip J, Bekele Y, Martins MV, Lund O, Franken KL, Khadge S, de Pontes MA, de Gonçalves HS (2012) Mycobacterium leprae virulence-associated peptides are indicators of exposure to M. leprae in Brazil, Ethiopia and Nepal. Mem Inst Oswaldo Cruz 107:112–123
Cheng X, Sun L, Zhao Q, Mi Z, Yu G, Wang Z, Sun Y, Wang C, Man C, Fu F, Liu H, Zhang F (2019) Development and evaluation of a droplet digital PCR assay for the diagnosis of paucibacillary leprosy in skin biopsy specimens. PLoS Negl Trop Dis 13:e0007284. https://doi.org/10.1371/journal.pntd.0007284
Cole ST, Eiglmeier K, Parkhill J, James KD, Thomson NR, Wheeler PR, Honoré N, Garnier T, Churcher C, Harris D, Mungall K, Basham D, Brown D, Chillingworth T, Connor R, Davies RM, Devlin K, Duthoy S, Feltwell T, Fraser A, Hamlin N, Holroyd S, Hornsby T, Jagels K, Lacroix C, Maclean J, Moule S, Murphy L, Oliver K, Quail MA, Rajandream MA, Rutherford KM, Rutter S, Seeger K, Simon S, Simmonds M, Skelton J, Squares R, Squares S, Stevens K, Taylor K, Whitehead S, Woodward JR, Barrell BG (2001) Massive gene decay in the leprosy bacillus. Nature 409:1007–1011
Corstjens PLAM, van Hooij A, Tjon Kon Fat EM, Alam K, Vrolijk LB, Dlamini S, da Silva MB, Spencer JS, Salgado CG, Richardus JH, van Hees CLM, Geluk A (2019) Fingerstick test quantifying humoral and cellular biomarkers indicative for M. leprae infection. Clin Biochem 66:76–82
de Souza-Santana FC, Marcos EVC, Nogueira MES, Ura S, Tomimori J (2015) Human leukocyte antigen class I and class II alleles are associated with susceptibility and resistance in borderline leprosy patients from Southeast Brazil. BMC Infect Dis 15(1):22
Deval H, Katoch K, Chauhan DS, Tyagi AK, Gupta RK, Kamal R, Kumar A, Yadav VS, Katoch VM, Hussain T (2016) TlyA protein of Mycobacterium leprae: a probable bio-marker of active infection. Lepr Rev 87:501–515
Doytchinova IA, Flower DR (2007) VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform 8:4. https://doi.org/10.1186/1471-2105-8-4
Duthie MS, Goto W, Ireton GC, Reece ST, Cardoso LPV, Martelli CMT, Stefani MMA, Nakatani M, de Jesus RC, Netto EM, Balagon MVF, Tan E, Gelber RH, Maeda Y, Makino M, Hoft D, Reed SG (2007) Use of protein antigens for early serological diagnosis of leprosy. Clin Vaccine Immunol CVI 14:1400–1408. https://doi.org/10.1128/cvi.00299-07
Duthie MS, Balagon MF, Maghanoy A, Orcullo FM, Cang M, Dias RF, Collovati M, Reed SG (2014) Rapid quantitative serological test for detection of infection with Mycobacterium leprae, the causative agent of leprosy. J Clin Microbiol 52:613–619. https://doi.org/10.1128/jcm.02085-13
Frade MAC, de Paula NA, Gomes CM, Vernal S, Bernardes Filho F, Lugão HB, de Abreu MMM, Botini P, Duthie MS, Spencer JS, Soares RCFR, Foss NT (2017) Unexpectedly high leprosy seroprevalence detected using a random surveillance strategy in midwestern Brazil: a comparison of ELISA and a rapid diagnostic test. PLoS Negl Trop Dis 11:e0005375. https://doi.org/10.1371/journal.pntd.0005375
Futami J, Atago Y, Azuma A, Putranto EW, Kinoshita R, Murata H, Sakaguchi M (2016) An efficient method for the preparation of preferentially heterodimerized recombinant S100A8/A9 coexpressed in Escherichia coli. Biochem Biophys Rep 6:94–100. https://doi.org/10.1016/j.bbrep.2016.03.009
Gasteiger E, Hoogland C, Gattiker A, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the ExPASy server. The proteomics protocols handbook. Humana Press, Totowa, pp 571–607
Geluk A, Duthie MS, Spencer JS (2011) Postgenomic Mycobacterium leprae antigens for cellular and serological diagnosis of M. leprae exposure, infection and leprosy disease. Lepr Rev 82(4):402
Hungria EM, de Oliveira RM, de Souza ALOM, Costa MB, de Souza VNB, Silva EA, Moreno FRV, Nogueira MES, Costa MRSN, Silva SMUR, Bührer-Sékula S, Reed SG, Duthie MS, de Stefani MM (2012) Seroreactivity to new Mycobacterium leprae protein antigens in different leprosy-endemic regions in Brazil. Memórias do Instituto Oswaldo Cruz 107:104–111. https://doi.org/10.1590/S0074-02762012000900017
Hungria EM, Bührer-Sékula S, de Oliveira RM, Aderaldo LC, de Pontes A, Cruz R, de Sa Gonçalves H, Penna MLF, Penna GO, de Stefani MM (2017) Leprosy reactions: the predictive value of Mycobacterium leprae-specific serology evaluated in a Brazilian cohort of leprosy patients (U-MDT/CT-BR). PLoS Negl Trop Dis. https://doi.org/10.1371/journal.pntd.0005396
Källberg M, Wang H, Wang S, Peng J, Wang Z, Lu H, Xu J (2012) Template-based protein structure modeling using the RaptorX web server. Nat Protoc 7:1511–1522
Kim HJ, Prithiviraj K, Groathouse N, Brennan PJ, Spencer JS (2013) Gene expression profile and immunological evaluation of unique hypothetical unknown proteins of Mycobacterium leprae by using quantitative real-time PCR. Clin Vaccine Immunol CVI 20:181–190. https://doi.org/10.1128/cvi.00419-12
Kumar A, Parkash O, Girdhar BK (2014) Analysis of antigens of Mycobacterium leprae by interaction to sera IgG, IgM, and IgA response to improve diagnosis of leprosy. Biomed Res Int 2014:283278
Leturiondo AL, Noronha AB, do Nascimento MOO, de Ferreira C, da Rodrigues F, Moraes MO, Talhari C (2019) Performance of serological tests PGL1 and NDO-LID in the diagnosis of leprosy in a reference Center in Brazil. BMC Infect Dis. https://doi.org/10.1186/s12879-018-3653-0
Lima FR, Takenami I, Cavalcanti MA, Riley LW, Arruda S (2017) ELISA-based assay of immunoglobulin G antibodies against mammalian cell entry 1A (Mce1A) protein: a novel diagnostic approach for leprosy. Mem Inst Oswaldo Cruz 112:844–849. https://doi.org/10.1590/0074-02760160549
Martinez AN, Talhari C, Moraes MO, Talhari S (2014) PCR-based techniques for leprosy diagnosis: from the laboratory to the clinic. PLoS Negl Trop Dis. https://doi.org/10.1371/journal.pntd.0002655
Meeker HC, Levis WR, Sersen E, Schuller-Levis G, Brennan PJ, Buchanan TM (1986) ELISA detection of IgM antibodies against phenolic glycolipid-I in the management of leprosy: a comparison between laboratories. Int J Lepr Other Mycobact Dis 54:530–539
Nagai H, Nukaga Y, Saeki K, Akabayashi A (2009) Self-regulation of recombinant DNA technology in Japan in the 1970s. Hist Sci Tokyo 19:1–18
Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786
Porto AC, Figueira RB, Barreto JA, Lauris JR (2015) Evaluation of the social, clinical and laboratorial profile of patients diagnosed with leprosy in a reference center in São Paulo. Bras Dermatol 90:169–177
Ramos CRR, Abreu PA, Nascimento ALTO, Ho PL (2004) A high-copy T7 Escherichia coli expression vector for the production of recombinant proteins with a minimal N-terminal His-tagged fusion peptide. Braz J Med Biol Res Rev Bras Pesqui Medicas E Biol 37:1103–1109. https://doi.org/10.1590/S0100-879X2004000800001
Reece ST, Ireton G, Mohamath R, Guderian J, Goto W, Gelber R, Groathouse N, Spencer J, Brennan P, Reed SG (2006) ML0405 and ML2331 are antigens of Mycobacterium leprae with potential for diagnosis of leprosy. Clin Vaccine Immunol 13:333–340
Reibel F, Cambau E, Aubry A (2015) Update on the epidemiology, diagnosis, and treatment of leprosy. Méd Mal Infect 45:383–393. https://doi.org/10.1016/j.medmal.2015.09.002
Richardus RA, van der Zwet K, van Hooij A, Wilson L, Oskam L, Faber R, van den Eeden SJF, Pahan D, Alam K, Richardus JH, Geluk A (2017) Longitudinal assessment of anti-PGL-I serology in contacts of leprosy patients in Bangladesh. PLoS Negl Trop Dis. https://doi.org/10.1371/journal.pntd.0006083
Roche PW, Failbus SS, Britton WJ, Cole R (1999) Rapid method for diagnosis of leprosy by measurements of antibodies to the M. leprae 35-kDa protein: comparison with PGL-I antibodies detected by ELISA and ”dipstick” methods. Int J Lepr Other Mycobact Dis 67:279–286
Saini C, Ramesh V, Nath I (2013) CD4+ Th17 cells discriminate clinical types and constitute a third subset of Non Th1, Non Th2 T cells in human leprosy. PLoS Negl Trop Dis. https://doi.org/10.1371/journal.pntd.0002338
Sampaio LH, Stefani MM, Oliveira RM, Sousa AL, Ireton GC, Reed SG, Duthie MS (2011) Immunologically reactive M. leprae antigens with relevance to diagnosis and vaccine development. BMC Infect Dis 11:26. https://doi.org/10.1186/1471-2334-11-26
Shankarkumar U, Ghosh K, Badakere S, Mohanty D (2003) Novel HLA Class I alleles associated with Indian leprosy patients. J Biomed Biotechnol 2003:208–211. https://doi.org/10.1155/S1110724303210019
Shen H-B, Chou K-C (2010) Gneg-mPLoc: a top-down strategy to enhance the quality of predicting subcellular localization of Gram-negative bacterial proteins. J Theor Biol 264:326–333. https://doi.org/10.1016/j.jtbi.2010.01.018
Simionatto S, Marchioro SB, Galli V, Hartwig DD, Carlessi RM, Munari FM, Laurino JP, Conceição FR, Dellagostin OA (2010) Cloning and purification of recombinant proteins of Mycoplasma hyopneumoniae expressed in Escherichia coli. Protein Expr Purif 69:132–136
Spencer JS, Kim HJ, Wheat WH, Chatterjee D, Balagon MV, Cellona RV, Tan EV, Gelber R, Saunderson P, Duthie MS, Reece ST, Burman W, Belknap R, Mac Kenzie WR, Geluk A, Oskam L, Dockrell HM, Brennan PJ (2011) Analysis of antibody responses to Mycobacterium leprae phenolic glycolipid I, lipoarabinomannan, and recombinant proteins To define disease subtype-specific antigenic profiles in leprosy. Clin Vaccine Immunol 18:260–267. https://doi.org/10.1128/cvi.00472-10
van Beers S, Hatta M, Klatser PR (1999) Seroprevalence rates of antibodies to phenolic glycolipid-I among school children as an indicator of leprosy endemicity. Int J Lepr Other Mycobact Dis 67:243–249
van Hooij A, Tjon Kon Fat EM, van den Eeden SJF, Wilson L, Batista da Silva M, Salgado CG, Spencer JS, Corstjens PLAM, Geluk A (2017) Field-friendly serological tests for determination of M. leprae-specific antibodies. Sci Rep. https://doi.org/10.1038/s41598-017-07803-7
van Hooij A, Tjon Kon Fat EM, Batista da Silva M, Carvalho Bouth R, Cunha Messias AC, Gobbo AR, Lema T, Bobosha K, Li J, Weng X, Salgado CG, Spencer JS, Corstjens PLAM, Geluk A (2018) Evaluation of immunodiagnostic tests for leprosy in Brazil China and Ethiopia. Sci Rep. https://doi.org/10.1038/s41598-018-36323-1
van Hooij A, van den Eden S, Richardus R, Fat ETK, Wilson L, Franken KLMC, Faber R, Khatun M, Alam K, Chowdhury AS, Richardus JH, Corstjens P, Geluk A (2019) Application of new host biomarker profiles in quantitative point-of-care tests facilitates leprosy diagnosis in the field. EBioMedicine 47:301–308. https://doi.org/10.1016/j.ebiom.2019.08.009
Vieira MCA, Nery JS, Paixão ES, Freitas de Andrade KV, Oliveira Penna G, Teixeira MG (2018) Leprosy in children under 15 years of age in Brazil: a systematic review of the literature. PLoS Negl Trop Dis. https://doi.org/10.1371/journal.pntd.0006788
WHO (2016) Global Leprosy Strategy 2016–2020: Accelerating towards a leprosy-free world
Yu C-S, Lin C-J, Hwang J-K (2004) Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci Publ Protein Soc 13:1402–1406. https://doi.org/10.1110/ps.03479604
Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FSL (2010) PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615. https://doi.org/10.1093/bioinformatics/btq249
Acknowledgements
We would like to thank the Center of Technological Development, Biotechnology (CDTEC) of the Federal University of Pelotas (UFPel) that has given us the expression strains.
Funding
This study was funded by Support Foundation for the Development of Education, Science and Technology in the State of Mato Grosso do Sul- FUNDECT (Grant Number 092/2015) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (Scholarship).
Author information
Authors and Affiliations
Contributions
Conceptualization, MSB and SBM; Methodology, MSB and IBAS; Formal analysis and investigation, MSB and IBAS; Writing—original draft preparation, MSB and SBM; Writing—review and editing, SB, SS and SBM; Funding acquisition, SBM and SS; Supervision, SB and SS. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
This study was approved by Research Ethical Committee (1.816.093-11/09/2016) of the Federal University of Grande Dourados (UFGD). All the participants signed an informed consent before enrolment.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Barbosa, M.d., de Sousa, I.B.A., Simionatto, S. et al. Recombinant polypeptide of Mycobacterium leprae as a potential tool for serological detection of leprosy. AMB Expr 9, 201 (2019). https://doi.org/10.1186/s13568-019-0928-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13568-019-0928-9