Abstract
Background
The Decipher 22-gene genomic classifier (GC) may help in post-radical prostatectomy (RP) decision making given its superior prognostic performance over clinicopathologic variables alone. However, most studies evaluating the GC have had a modest representation of African-American men (AAM). We evaluated the GC within a large Veteran Affairs cohort and compared its performance to CAPRA-S for predicting outcomes in AAM and non-AAM after RP.
Methods
GC scores were generated for 548 prostate cancer (PC) patients, who underwent RP at the Durham Veteran Affairs Medical Center between 1989 and 2016. This was a clinically high-risk cohort and was selected to have either pT3a, positive margins, seminal vesicle invasion, or received post-RP radiotherapy. Multivariable Cox models and survival C-indices were used to compare the performance of GC and CAPRA-S for predicting the risk of metastasis and PC-specific mortality (PCSM).
Results
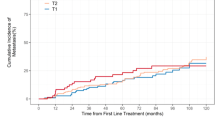
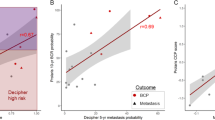
Median follow-up was 9 years, during which 37 developed metastasis and 20 died from PC. Overall, 55% (n = 301) of patients were AAM. In multivariable analyses, GC (high vs. intermediate and intermediate vs. low) was a significant predictor of metastasis in all men (all p < 0.001). Consistent with prior studies, relative to CAPRA-S, GC had a higher C-index for 5-year metastasis (0.78 vs. 0.72) and 10-year PCSM (0.85 vs. 0.81). There was a suggestion GC was a stronger predictor in AAM than non-AAM. Specifically, the 5-year metastasis risk C-index was 0.86 in AAM vs. 0.69 in non-AAM and the 10-year PCSM risk C-index was 0.91 in AAM vs. 0.78 in non-AAM. However, the test for interaction of race and the performance of the GC in the Cox model was not significant for either metastasis or PCSM (both p ≥ 0.3).
Conclusions
GC was a very strong predictor of poor outcome and performed well in both AAM and non-AAM. Our data support the use of GC for risk stratification in AAM post-RP. While our data suggest that GC may actually work better in AAM, given the limited number of events, further validation is needed.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 4 print issues and online access
$259.00 per year
only $64.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Sanda MG, Cadeddu JA, Kirkby E, Chen RC, Crispino T, Fontanarosa J, et al. Clinically localized prostate cancer: AUA/ASTRO/SUO guideline. Part II: recommended approaches and details of specific care options. J Urol. 2018;199:990–7.
Cooperberg MR, Hilton JF, Carroll PR. The CAPRA-S score: a straightforward tool for improved prediction of outcomes after radical prostatectomy. Cancer. 2011;117:5039–46.
Ross AE, D'Amico AV, Freedland SJ. Which, when and why? Rational use of tissue-based molecular testing in localized prostate cancer. Prostate Cancer Prostatic Dis. 2016;19:1–6.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30.
Gaines AR, Turner EL, Moorman PG, Freedland SJ, Keto CJ, McPhail ME, et al. The association between race and prostate cancer risk on initial biopsy in an equal access, multiethnic cohort. Cancer Causes Control. 2014;25:1029–35.
Chu DI, Moreira DM, Gerber L, Presti JC Jr., Aronson WJ, Terris MK, et al. Effect of race and socioeconomic status on surgical margins and biochemical outcomes in an equal-access health care setting: results from the Shared Equal Access Regional Cancer Hospital (SEARCH) database. Cancer. 2012;118:4999–5007.
Wallace TA, Prueitt RL, Yi M, Howe TM, Gillespie JW, Yfantis HG, et al. Tumor immunobiological differences in prostate cancer between African-American and European-American men. Cancer Res. 2008;68:927–36.
Kim HS, Moreira DM, Jayachandran J, Gerber L, Banez LL, Vollmer RT, et al. Prostate biopsies from black men express higher levels of aggressive disease biomarkers than prostate biopsies from white men. Prostate Cancer Prostatic Dis. 2011;14:262–5.
Vijayakumar S, Henegan JC, Zhang X, Wang W, Day WA, Vijayakumar V, et al. Enriching gene expression profiles will help personalize prostate cancer management for African-Americans: a perspective. Urologic Oncol. 2017;35:315–21.
Ross AE, Den RB, Yousefi K, Trock BJ, Tosoian J, Davicioni E, et al. Efficacy of post-operative radiation in a prostatectomy cohort adjusted for clinical and genomic risk. Prostate Cancer Prostatic Dis. 2016;19:277–82.
Spratt DE, Yousefi K, Deheshi S, Ross AE, Den RB, Schaeffer EM, et al. Individual patient-level meta-analysis of the performance of the decipher genomic classifier in high-risk men after prostatectomy to predict development of metastatic disease. J Clin Oncol. 2017;35:1991–8.
Spratt DE, Dai DLY, Den RB, Troncoso P, Yousefi K, Ross AE, et al. Performance of a prostate cancer genomic classifier in predicting metastasis in men with prostate-specific antigen persistence postprostatectomy. Eur Urol. 2018;74:107–14.
McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. REporting recommendations for tumour MARKer prognostic studies (REMARK). Br J Cancer. 2005;93:387–91.
Pepe MS, Feng Z, Janes H, Bossuyt PM, Potter JD. Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: standards for study design. J Natl Cancer Inst. 2008;100:1432–8.
Den RB, Yousefi K, Trabulsi EJ, Abdollah F, Choeurng V, Feng FY, et al. Genomic classifier identifies men with adverse pathology after radical prostatectomy who benefit from adjuvant radiation therapy. J Clin Oncol. 2015;33:944–51.
Lockstone HE. Exon array data analysis using Affymetrix power tools and R statistical software. Brief Bioinform. 2011;12:634–44.
Piccolo SR, Sun Y, Campbell JD, Lenburg ME, Bild AH, Johnson WE. A single-sample microarray normalization method to facilitate personalized-medicine workflows. Genomics. 2012;100:337–44.
Erho N, Crisan A, Vergara IA, Mitra AP, Ghadessi M, Buerki C, et al. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PloS ONE. 2013;8:e66855.
Karnes RJ, Bergstralh EJ, Davicioni E, Ghadessi M, Buerki C, Mitra AP, et al. Validation of a genomic classifier that predicts metastasis following radical prostatectomy in an at risk patient population. J Urol. 2013;190:2047–53.
Ross AE, Johnson MH, Yousefi K, Davicioni E, Netto GJ, Marchionni L, et al. Tissue-based genomics augments post-prostatectomy risk stratification in a natural history cohort of intermediate- and high-risk men. Eur Urol. 2016;69:157–65.
Briganti A, Karnes RJ, Joniau S, Boorjian SA, Cozzarini C, Gandaglia G, et al. Prediction of outcome following early salvage radiotherapy among patients with biochemical recurrence after radical prostatectomy. Eur Urol. 2014;66:479–86.
Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56:337–44.
Gray RJ. A class of K-sample tests for comparing the cumulative incidence of a competing risk. Ann Stat. 1988;16:1141–54.
Nguyen PL, Haddad Z, Ross AE, Martin NE, Deheshi S, Lam LLC, et al. Ability of a genomic classifier to predict metastasis and prostate cancer-specific mortality after radiation or surgery based on needle biopsy specimens. Eur Urol. 2017;72:845–52.
Canter DJ, Freedland S, Rajamani S, Latsis M, Variano M, Halat S, et al. Analysis of the prognostic utility of the cell cycle progression (CCP) score generated from needle biopsy in men treated with definitive therapy. Prostate Cancer Prostatic Dis. 2019.
Koch MO, Cho JS, Kaimakliotis HZ, Cheng L, Sangale Z, Brawer M, et al. Use of the cell cycle progression (CCP) score for predicting systemic disease and response to radiation of biochemical recurrence. Cancer Biomark. 2016;17:83–8.
Van Den Eeden SK, Lu R, Zhang N, Quesenberry CP Jr., Shan J, Han JS, et al. A biopsy-based 17-gene genomic prostate score as a predictor of metastases and prostate cancer death in surgically treated men with clinically localized disease. Eur Urol. 2018;73:129–38.
Cedars BE, Washington SL, 3rd, Cowan JE, Leapman M, Tenggara I, Chan JM, et al. Stability of a 17-gene genomic prostate score in serial testing of men on active surveillance of early stage prostate cancer. J Urol. 2019: 101097ju0000000000000271.
Cullen J, Rosner IL, Brand TC, Zhang N, Tsiatis AC, Moncur J, et al. A biopsy-based 17-gene genomic prostate score predicts recurrence after radical prostatectomy and adverse surgical pathology in a racially diverse population of men with clinically low- and intermediate-risk prostate cancer. Eur Urol. 2015;68:123–31.
Freedland SJ, Gerber L, Reid J, Welbourn W, Tikishvili E, Park J, et al. Prognostic utility of cell cycle progression score in men with prostate cancer after primary external beam radiation therapy. Int J Radiat Oncol Biol Phys. 2013;86:848–53.
Cullen J, Young D, Chen Y, Degon M, Farrell J, Sedarsky J, et al. Predicting prostate cancer progression as a function of ETS-related gene status, race, and obesity in a longitudinal patient cohort. Eur Urol Focus. 2018;4:818–24.
You S, Knudsen BS, Erho N, Alshalalfa M, Takhar M, Al-Deen Ashab H, et al. Integrated classification of prostate cancer reveals a novel luminal subtype with poor outcome. Cancer Res. 2016;76:4948–58.
Zhao SG, Chang SL, Erho N, Yu M, Lehrer J, Alshalalfa M, et al. Associations of luminal and basal subtyping of prostate cancer with prognosis and response to androgen deprivation therapy. JAMA Oncol. 2017;3:1663–72.
Tomlins SA, Alshalalfa M, Davicioni E, Erho N, Yousefi K, Zhao S, et al. Characterization of 1577 primary prostate cancers reveals novel biological and clinicopathologic insights into molecular subtypes. Eur Urol. 2015;68:555–67.
Faisal FA, Sundi D, Tosoian JJ, Choeurng V, Alshalalfa M, Ross AE, et al. Racial variations in prostate cancer molecular subtypes and androgen receptor signaling reflect anatomic tumor location. Eur Urol. 2016;70:14–7.
Powell IJ, Dyson G, Land S, Ruterbusch J, Bock CH, Lenk S, et al. Genes associated with prostate cancer are differentially expressed in African American and European American men. Cancer Epidemiol Biomark Prev. 2013;22:891–7.
Yamoah K, Johnson MH, Choeurng V, Faisal FA, Yousefi K, Haddad Z, et al. Novel biomarker signature that may predict aggressive disease in African American men with prostate cancer. J Clin Oncol. 2015;33:2789–96.
Acknowledgements
This study was funded by Decipher Biosciences.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
JZ, NF, and ED are employees of Decipher Biosciences.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Howard, L.E., Zhang, J., Fishbane, N. et al. Validation of a genomic classifier for prediction of metastasis and prostate cancer-specific mortality in African-American men following radical prostatectomy in an equal access healthcare setting. Prostate Cancer Prostatic Dis 23, 419–428 (2020). https://doi.org/10.1038/s41391-019-0197-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41391-019-0197-3
This article is cited by
-
Genomic classifiers and prognosis of localized prostate cancer: a systematic review
Prostate Cancer and Prostatic Diseases (2024)
-
Disparities in prostate cancer diagnosis and management: recognizing that disparities exist at all junctures along the prostate cancer journey
Prostate Cancer and Prostatic Diseases (2023)
-
SR9009 inhibits lethal prostate cancer subtype 1 by regulating the LXRα/FOXM1 pathway independently of REV-ERBs
Cell Death & Disease (2022)
-
Salvage therapy for prostate cancer after radical prostatectomy
Nature Reviews Urology (2021)
-
A comparative study of PCS and PAM50 prostate cancer classification schemes
Prostate Cancer and Prostatic Diseases (2021)