Abstract
The functional role of expansin-like A and B family members remains unclear in plants. In this study, we investigate the functional role of Brassica rapa expansin-like B1 (BrEXLB1) by overexpressing BrEXLB1 sense, antisense and BrEXLB1 specific promoter: GUS in Arabidopsis. The overexpressors of sense BrEXLB1, antisense BrEXLB1 showed variable and unstable pleiotropic effects on leaf growth. Interestingly, overexpressors of sense BrEXLB1 enhances plant height than antisense overexpressors and wild types. GUS reporter-aided analysis of BrEXLB1 promoter showed their activity prominently in seeds, roots and carpels. This is further confirmed by relative quantification of BrEXLB1 among various tissue samples of B. rapa by semi-quantitative Reverse transcription-polymerase chain reaction assay (RT-PCR). We found that BrEXLB1 promoter has several cis-acting elements including light-responsive, phytohormone-responsive, environmental/stress-responsive and tissue-specific elements in promoter sequences. Hence, we analysed the promoter activity of BrEXLB1 by GUS reporter-aided approach, in which stress, phytohormones and other factors regulated changes in GUS transcripts were measured using quantitative RT-PCR. This study suggests that promoter activity is inducible to exogenous application of phytohormones such as indole-3-acetic acid, jasmonic acid, and other factors such as white light and drought stress. These results suggest that BrEXLB1 under its specific promoters may participate in regulation of leaf, plant growth and responds to hormone availability, light quality, dark periods, developmental stages, and drought conditions.
Similar content being viewed by others
Introduction
Expansins are cell wall proteins, present in all plants, play crucial role in cell wall remodeling (Marowa et al. 2016). Plant expansins are necessary for growth and development as it helps the plants to uptake water, cell stress relaxation and or cell enlargement through its cell wall loosening activity (Cosgrove 2001; Sasidharan et al. 2011). It is a non-enzymatic proteins that extend cell walls by weakening non-covalent linkages between cellulose microfibrils and xyloglucans (Santiago et al. 2018; Wang et al. 2013). Based on the phylogenetic relationships, plant expansins are classified into four families: α-expansin (EXPA), β-expansin (EXPB), expansin-like A (EXLA), and expansin-like B (EXLB) proteins (Kende et al. 2004). Members of EXPA and EXPB families have been clearly shown to be involved in most plant growth and developmental processes, whereas EXLA and EXLB proteins are only known from their homologous conserved gene sequences and their functions yet to be discovered (Cosgrove 2015).
Following initial investigations of expansins in cucumber hypocotyls (McQueen-Mason and Cosgrove 1994; McQueen-Mason et al. 1992), numerous studies have shown that expansins are involved in all cell wall modification processes, from abscission (Cho and Cosgrove 2000), seed number increase (Bae et al. 2014), root formation (Greenwood et al. 2006), fruit ripening (Brummell et al. 1999), plant architecture definition (Dal Santo et al. 2011) to plant–microbe interactions (Wieczorek et al. 2006). Although the exact mechanism underlying expansin activity is not fully understood (Cosgrove 2015), several studies have successfully correlated the involvement of expansins with leaf growth and development using sense and antisense transgenic plants. Examples in Arabidopsis include overexpression of NtEXPA5 increasing leaf and stem sizes (Kuluev et al. 2013), sense and antisense overexpression of AtEXPA10 effectively altering leaf and petiole sizes (Cho and Cosgrove 2000), and overexpression of AtEXPA10 and PnEXPA1 producing larger leaves and longer stems (Kuluev et al. 2012). Overexpression of NtEXPA1 increases tobacco leaf size (Kuluev et al. 2014), and PttEXPA1 overexpression increases stem internode elongation and leaf expansion in aspen (Gray-Mitsumune et al. 2008). Furthermore, CsEXP1 overexpression has been found to reduce leaf sizes and internodes in tomato plants (Wieczorek et al. 2006). Overexpression of OsEXP4 sense and antisense constructs significantly increases and decreases, respectively, the number of leaves in rice plants (Choi et al. 2003). Because EXLA and EXLB gene sequences are conserved across many plant species and are highly homologous to those of EXPA and EXPB members (Sampedro and Cosgrove 2005), EXLA and EXLB genes may be directly and/or indirectly involved in plant growth and developmental processes. Concurrence with this hypothesis, Arabidopsis EXLA2 gene has been recently reported to involve in plant development and defense (Abuqamar et al. 2013).
In our previous comparative genomic analysis, we identified three EXLB genes in the Chinese cabbage (Brassica rapa subsp. pekinensis) genome (Krishnamurthy et al. 2015). In this study, we cloned and characterized the physiological effect of the BrEXLB1 gene in transgenic Arabidopsis expressing BrEXLB1 sense and antisense constructs driven by the CaMV35S promoter. We also isolated the BrEXLB1 promoter sequence and characterized its activity under various environmental stimuli, phytohormones, and stress conditions. The promoter activity was effectively measured using reporter gene expression levels in transgenic Arabidopsis via quantitative reverse-transcription PCR (qRT-PCR). This study will help to understand the likely function of BrEXLB1 in plant growth and development.
Materials and methods
Sequence analysis
Full-length coding, amino acid, and upstream (promoter region: − 1600 bp to + 1 bp) sequences of EXLB genes of B. rapa (BrEXLB1) and Arabidopsis thaliana (AtEXLB1) were retrieved from Brassica (http://brassicadb.org/brad/index.php; B. rapa genome version 1.5) and Phytozome v10.3 (http://phytozome.jgi.doe.gov/ pz/portal.html#) databases, respectively. To identify putative cis-acting elements of BrEXLB, the promoter region (− 1500 bp to + 1 bp) was searched against known motifs of plantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) database. All tools and programs mentioned in this study were used with default settings unless otherwise specified.
Construction of expression cassette for BrEXLB1-sense, antisense and BrEXLB1 promoter::GUS
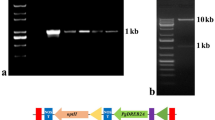
The BrEXLB1 coding sequence was amplified by polymerase chain reaction (PCR) with gene-specific forward (5′-AATATGAAGACATTTAACGTCTTG-3′) and reverse (5′-GGAATCAAGTAAGTAGAATGTTGG-3′) primers. The EcoR I digested PCR amplicons (0.773 kb) were inserted in the transgene orientation between the cauliflower mosaic virus 35S promoter (CaMV35Sp) and the nopaline synthase terminator of a pCAMBIA1390 vector to generate binary vectors pCAMBIA1390::35S-Pro + BrEXLB1sense (pLSI09) and pCAMBIA1390::35S-Pro + BrEXLB1antisense (pLSI12). The sense and the antisense orientation of BrEXLB1 was confirmed by Sanger sequencing. Similarly, the BrEXLB1 promoter region was PCR amplified using forward and reverse primers 5′-GAAACGAACACGGCTATTATACG-3′ and 5′-CATATTGTTATATGTAACTATTGTA-3′, respectively. The amplified promoter sequence (1.5 kb) was ligated into a pGEM-T Easy vector followed by transformation into Escherichia coli DH-5α cells for amplification, and then introduced into a pCAMBIA1391Z vector at EcoR I site to generate the binary vector pCAMBIA1391Z + BrEXLB1promoter::GUS (pLSI13). All primer pairs were obtained from GenoTech (Daejeon, Korea).
Plant materials, Agrobacterium mediated transformation and morphology of transgenic lines
Arabidopsis thaliana Columbia (Col-0) seeds were surface-sterilized by 70% ethanol for 15 min followed by 100% ethanol for 2 min and plated on Murashige and Skoog (MS) medium supplemented with vitamins, 1.5% sucrose, and 0.25% phytagel. Plates were stratified at 4 °C under dark for 2 days to induce synchronous germination and transferred to a growth chamber (16-h light/8-h dark photoperiod at 23 °C) for transformation and morphological character analyses. Transformation (A. tumefaciens strain GV3101) and transformant selection with hygromycin B were performed according to Hong et al. (2012). Leaf length, leaf width, and petiole length were measured on 15, 20, and 30 days after transferring the pots into the growth chamber. Measurements were taken from three independent biological replicates. Mean significant differences were compared by Duncan Multiple Range Test at P ≤ 0.05 using SAS package 9.1.3 service pack 4.
Histochemical localization of β-glucuronidase (GUS) activity
To evaluate the regulation of BrEXLB1 promoter under different developmental stages, seeds of transgenic Arabidopsis (pLSI13) lines were germinated as mentioned above. Samples were collected from the first day of germination, 7-, 14-, and 20-days old seedlings and reproductive organs of mature plants. Samples were directly used for the GUS assay. An assay was performed using a β-Glucuronidase Reporter Gene staining kit (Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer’s instructions.
Phytohormone, stress treatments, and quantitative reverse-transcription real-time PCR (qRT-PCR)
Transgenic T1 Arabidopsis seedlings harboring pLSI13 cassette were surface sterilized as mentioned above and plated on MS medium supplemented with vitamins, 1.5% sucrose, 0.25% phytagel and 30 μg/ml hygromycin B. After 2 days stratification under dark at 4 °C and 5 days under growth chamber, transformants were selected based on the hypocotyl length (Harrison et al. 2006). Selected seedlings were divided for triplicate and independently grown hydroponically in MS solution for 3 days in Arabidopsis growth chamber. Then, three plates each (each having three seedlings) were moved to different light qualities namely white (photosynthetic photo flux density: 59 μmol m−2 s−1), blue (59 μmol m−2 s−1), red (10 μmol m−2 s−1), far-red (2 μmol m−2 s−1) and dark in our customized LED chamber. Concurrently, phytohormones, namely IAA (50 μM), GA (50 μM), JA (100 μM), SA (100 μM), and ABA (100 μM), as well as heavy metal (CdCl2; 200 μM) and polyethylene glycol (PEG6000; 4%) were added to the respective triplicate hydroponic cultures and kept in the growth chamber with gentle shaking (60 rpm). After incubation for 6 h, entire seedlings from all the treatments were harvested, frozen in liquid nitrogen, and stored at − 80 °C. Seedlings grown in growth chamber (80 μmol m−2 s−1) in MS solution were used as control. Temperature (25 ± 0.5 °C) and humidity (33 ± 2%) were maintained same in both the growth chamber and LED chamber.
Total RNA was extracted using Ambion Purelink® RNA Mini Kit (Thermo Fisher Scientific, MA, USA) from ground tissues samples (Qiagen TissueLyser II (Hilden, Germany). RNA (1 µg) was reverse transcribed into cDNA using Qiagen`s QuantiTect® Reverse Transcription Kit (Hilden, Germany) which included genomic DNA elimination step. cDNA products were diluted in a 1:9 ratio with nuclease-free water (Promega) and used as template for qRT-PCR. QRT-PCR was performed using the SYBR Premix Ex Taq kit® (TaKaRa, Japan) with GUS-specific primers (forward: 5′-GAATACGGCGTGGATACGTTAG-3′ and reverse 5′-GATCAAAGACGCGGTGATACA-3′) on StepOnePlus Real-Time PCR System (Applied Biosystems, CA, USA) under the following cycling profile: 95 °C for 30 s, 95 °C for 5 s (40 cycles), 60 °C for 30 s, and melting curve analysis at 65 °C for 10 s with 61 cycles. The reaction mixture volume for qRT-PCR was 20 µL. Experimental repeat runs for three biological and three technical replicates were included in the analysis. The Arabidopsis Actin2 (AT3G18780) gene (forward primer: 5′-TCGGTGGTTCCATTCTTGCT-3′; reverse primer: 5′-GCTTTTTAAGCCTTT GATCTTGAGAG-3′) was used as an internal control to normalize the expression level of the target GUS gene. An analysis of the relative quantitation (RQ) values were directly inferred from the StepOnePlus Real-Time PCR System that mean significant differences were compared by Duncan Multiple Range Test at P ≤ 0.05 using SAS package 9.1.3 service pack 4.
Relative quantification of BrEXLB1 transcripts in different tissues of Brassica rapa
Brassica rapa ‘Chiifu-401-42’ seeds were germinated and grown according to Hong et al. (2010). Samples of apical meristems, cotyledons and hypocotyls were collected from 2-week-old B. rapa seedlings whereas leaves, roots, pollen, carpels, and siliques were harvested from mature B. rapa plants. All samples were frozen in liquid nitrogen and stored at − 80 °C throughout the sampling period and were ground using Qiagen TissueLyser II (Hilden, Germany). Total RNAs and cDNA were prepared as mentioned above and used as template for semi-quantitative (reaction volume 25 µL) PCR with BrEXLB1 primers under following cycling parameters: 95 °C for 5 min, followed by 25 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 45 s, with a final extension step of 72 °C for 7 min. B. rapa β-actin (forward: 5′-TGGCATCACACTTTTCTACAA-3′; reverse: 5′-CAACGGAATCTCTCAGCTCC-3′) was used as an internal control to check the RNA quality (Hong et al. 2010).
Results and discussion
Implications on the structural characteristics of B. rapa EXLB genes and their promoter regions
Of the four expansin families, EXPA is the largest, followed in order by EXPB, EXLB, and EXLA. The highest numbers of EXLA (n = 49) and EXLB (n = 15) genes have been found in the soybean genome whereas the highest number of EXPB (n = 48) and EXLA (n = 4) genes have been identified in the maize genome (Cosgrove 2015). The Arabidopsis genome contains 36 EXPA, 6 EXPB, 3 EXLA and 1 EXLB genes. The whole genome triplication event generated at least 53 expansin genes in the B. rapa genome that have occurred after the speciation of Brassica from Arabidopsis (Krishnamurthy et al. 2015). The B. rapa genome consists of three EXLB genes namely BrEXLB1 (gene id: Bra040107), BrEXLB2 (Bra012687), and BrEXLB3 (Bra012684) that were orthologous to AtEXLB1 (At4G17030) ((Krishnamurthy et al. 2015); Table S1). Coding sequences of all BrEXLBs were of similar length (753 bp). BrEXLB2 and BrEXLB3 were tandem duplicates that shared 95.9% nucleotide and 97.5% amino-acid sequence similarity, while nucleotide and amino acid sequences of BrEXLB1 shared ~ 90% similarity with those of BrEXLB1 and BrEXLB2. All three BrEXLB protein sequences comprised 250 amino acids with nearly identical molecular weights of 27.8 kDa. The isoelectric points of BrEXLB2 and BrEXLB3 were the same (6.51) and slightly different from that of BrEXLB1 (6.30). Multiple sequence alignment revealed that AtEXLB1 shared 88–90% sequence similarity at nucleotide and amino acid levels with BrEXLBs suggesting that AtEXLB1 and BrEXLBs may share common function. Further to understand evolutionary relationships between BrEXLB1 and homologs found in other closely related species of Brassicaceae families such as Brassica oleracea var. oleracea (BoEXLB1), Arabidopsis thaliana (AtEXLB1), Capsella rubella (CrEXLB1), Camelina sativa (CsEXLB1), Brassica napus (BnEXLB1) and Raphanus sativus (RsEXLB1) were compared through multiple sequence alignment followed by phylogenetic tree analysis (Fig. 1). BrEXLB1 was more similar to BoEXLB1 than BnEXLB1. The close relationship among EXLB1 of the species can be ranked as BoEXLB1 followed by BnEXLB1, RsEXLB1, AtEXLB1, CsEXLB1 and CrEXLB1. Interestingly, the similarity between BrEXLB1and RsEXLB1 is higher than AtEXLB1.
Phylogenetic relationship between Brassica rapa EXLB1 and homolog EXLB1 gene sequences of closely related species of Brassicaceae families. The proteins compared are Brassica rapa (BrEXLB1), Brassica oleracea var. oleracea (BoEXLB1), Arabidopsis thaliana (AtEXLB1), Capsella rubella (CrEXLB1), Camelina sativa (CsEXLB1), Brassica napus (BnEXLB1) and Raphanus sativus (RsEXLB1)
Comparative sequence alignment among the promoter regions (− 1600 bp to + 1 bp of the upstream sequence) of BrEXLBs revealed that the BrEXLB1 promoter share only 48–50% similarity to BrEXLB2 and BrEXLB3 promoters, whereas 72.31% similarity was found between the latter two. The AtEXLB1 promoter showed 43–49% similarity to the BrEXLB promoters. Due to the limitation of PlantCARE database, the − 1500 bp to + 1 bp of the obtained promoter sequences were submitted to identify and compare cis-acting elements. In each sequence, 83–96 conserved cis-acting elements were identified corresponding to 55 unique elements that could be broadly classified into five types: (1) promoter-related, (2) light-responsive, (3) plant hormone-responsive, (4) environmental- and stress-responsive, and (5) tissue-specific (Table S2). Of the 55 cis-acting elements, 35–47% is contributed by promoter-related elements followed by light-responsive (20–33%) and phytohormone-responsive elements. BrEXLB promoters has several elements like phytohormone responsive (ABA and SA), fungal elicitor-responsive (Box-W1 (TTGACC), wound and pathogen responsive (W box: (TTGACC)), meristem expression-specific (CAT-box: GCCACT), endosperm-specific expression (Skn-1_motif: GTCAT), circadian control and element involved in negative regulation of phloem expression and responsible for restricting vascular expression (AC-II; [(C/T)T(T/C)(C/T)(A/C)(A/C)C(A/C)A(A/C) C(C/A) (C/A)C]. The comparative analysis with BrEXLB2, BrEXLB3 and its orthologous promoter AtEXLB1 shows some elements are specific to respective promoter sequences. For instance, BrEXLB1 has unique elements like Box-W1 and the W box (TTGACC), similarly, BrEXLB2 has MBS motif (CAACTG; involved in drought inducibility), BrEXLB3 has 5′-untranslated region Py-rich stretch element (TTTCTTCTCT) and AtEXLB1 has auxin, JA-, GA-responsive, and AT-rich sequence (TAAAAATACT; element for maximal elicitor-mediated activation).
Elements such as anaerobic induction-responsive (ARE motif: TGGTTT), stress-responsive (TC-rich repeats: GTTTTCTTAC) were commonly found in all BrEXLB promoters while CAT-box was found in BrEXLB1 and BrEXLB3 promoters. Similarly heat stress-responsive elements (HSE motif: AAAAAATTTC and AGAAAATTCG) were present in BrEXLB2 and BrEXLB3 promoters. Further comparison with AtEXLB1 revealed that AC-II as well as the Skn-1_motif: GTCAT are common among all the promoters, whereas circadian control elements were unique to BrEXLB promoters. As indicated in the present study and by the reports of Zhu et al. (2014), we expect that the presence of diverse cis elements may facilitate the differential expression of EXLB genes depending upon the environmental stumuli, presence of phytohormones or its exogenous application, developmental stages and stress conditions.
GUS reporter-aided analysis of BrEXLB1-specific promoter activity during plant development, application of phytohormones and stress in Arabidopsis
To investigate how BrEXLB1 regulated in different tissues, a GUS reporter-aided approach was adopted to elucidate the promoter activities of BrEXLB1 in transgenic Arabidopsis in the present study. The identification of putative cis-acting elements using PlantCARE revealed that BrEXLBs has several elements including specific and common elements found in other BrEXP and Arabidopsis. Moreover, studies shown that abiotic stresses (Chen et al. 2018) including drought, light, phytohormones and hard metals regulates the expression of expansin (Marowa et al. 2016; Santiago et al. 2018). We thus generated transgenic Arabidopsis homozygous lines harboring the cassette pCAMBIA1391Z + BrEXLB1promoter::GUS (pLSI13) to assess the functional regulation of the BrEXLB1 promoter during developmental stages and in response to various light qualities, phytohormones, and drought stress. GUS activity was clearly detectable immediately after radicle elongation, on the first day of germination (Fig. 2a). Staining was observed in the hypocotyls, cotyledonary leaves, and radicles, with the strongest staining found in the apical points of 3- and 7-day-old seedlings (Fig. 2b, c). GUS staining was uniformly present in rosettes and hypocotyls but was unevenly detected in the root systems of 20-day-old seedlings (Fig. 2d). In addition, GUS activity was detected in pollen (Fig. 2e); activity was also detected in stigmas, with intense staining restricted to the abscission zone and stigmatic papillae with no expression in the middle part (Fig. 2f). These results indicate BrEXLB1 likely to play a role in development of apical points in early development stages, hypocotyls, cotyledonary leaves, radicles, abscission zone and stigmatic papillae and pollen and to some extent in root.
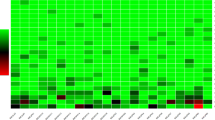
Further to test the importance of cis-acting elements of BrEXLB1 promoter under different light qualities, phytohormone and PEG induced drought stress was evaluated by quantitate the abundance of GUS transcript levels in each treatment using transgenic pLSI13 lines (Fig. 3). The results show that the transcript levels of GUS was 0.3-fold increased by white light and 0.3-0.4-fold decreased by red, far-red and dark conditions while no significant change was observed under blue light. It is therefore conceivable that light qualities and dark periods mediate plant growth through regulation of EXPLB1. The previous reports on EXPA and EXPB from various studies showed that phytohormones (auxin, gibberellin, cytokinin, and ethylene) regulate EXPA and EXPB gene expression at the transcript level (Cho 1997; Hutchison et al. 1999; Kim et al. 2000; Wrobel and Yoder 2001).
GUS expression in pBrEXLB1::GUS transgenic Arabidopsis by qRT-PCR. Ten-day-old transgenic seedlings were treated with different light qualities (white, blue, red, far-red and dark), phytohormones [indole-3-acetic acid (IAA), gibberellic acid (GA), jasmonic acid (JA), salicylic acid (SA), abscisic acid (ABA)], heavy metal (CdCl2), and drought (PEG) stress for 6 h. Arabidopsis Actin2 gene expression was used to normalize the expression of GUS. Values shown are the mean and SD of three independent biological and technical experiments. Asterisks indicate statistical significant differences from the control (Duncan Multiple Range Test at P ≤ 0.05) (color figure online)
Similarly, the exogenous application of IAA, JA, and exposure to PEG6000 mediated drought stress and CdCl2 influence the activities of BrEXLB1 promoter. The quantitative RT-PCR analysis showed that application of IAA increased GUS transcripts by 2.8-fold and, JA and PEG exhibited 2.5-fold increase in pLSI13 lines. Intriguingly, cis-elements responsive to IAA, JA- and drought stress were not observed from the sequence analysis (Table S2). However, both IAA (auxin), JA-responsive elements were found in AtEXLB1 which is a homolog of BrEXLB1. It is reasonable to think that the technical limitation for sequence length with PlantCARE might reduce our chance in identification of auxin-, JA-responsive elements. Moreover, standardization of time periods for each treatment may provide most conceivable data. Further, the synergistic relationship between JA- and wound-responsive elements (W-box) may facilitate the induced activity of BrEXLB1 promoter against JA (Bari and Jones 2009). Up-regulation by PEG6000 and slightly down-regulation by CdCl2 suggests BrEXLB1 is sensitive to plant water balance, as both CdCl2 and PEG6000 decrease plant water potential and thereby inhibit plant growth and development (Lagerwerff et al. 1961; Perfus-Barbeoch et al. 2002). As evident from the results (Fig. 3), BrEXLB1 promoter mediated expression of GUS transcript is also regulated by exogenous application of GA, SA, ABA and CdCl2, although their exposure did not significantly affect the promoter activity. PEG induced activity of BrEXLB1 are encouraging as it may result in improved drought stress response as noted in tobacco (Chen et al. 2016). Nevertheless, our results show that BrEXLB1 promoter activity is up-regulated by white light, IAA, JA and PEG treatments and reduced under red, far-red and dark treatments indicating the possible involvement of BrEXLB1 gene in plant growth and development.
Impact of BrEXLB1 sense and antisense overexpressing transgenic Arabidopsis on plant growth and development
Transcript levels of endogenous BrEXLB1 in B. rapa were examined using semi-quantitative RT-PCR. As inferred from the amplicon intensity, the expression of BrEXLB1 was high in seeds, roots, and carpels, less in apical meristems and hypocotyls, and undetectable in cotyledons, leaves, pollen, and siliques (Fig. 4a).
Relative quantification of BrEXLB1 across different tissues of B. rapa, Impact assessment in overexpressors of sense and antisense constructs on leaf growth in Arabidopsis. a Expression pattern of BrEXLB1 in different tissues (apical meristem, cotyledon, hypocotyl) of two-week old mature B. rapa ‘Chiifu-401-42’ plants (leaf, root, pollen, carpel, silique) analyzed by semi-quantitative RT-PCR. Relative quantification of BrEXLB1 in sense (b) and antisense (c) lines (whole-seedlings of two-week old transgenic Arabidopsis) by semi-quantitative RT-PCR. Blade length (d), blade width (e), and petiole length (f) in transgenic Arabidopsis lines were measured at 15, 20 and 30 days after transferring the pots into the growth chamber. Measurements shown are the mean and SD of three independent biological with three seedlings per line/wild type. Statistical significance between WT (wild type) and transgenic plants in each test at P ≤ 0.05 (*)
One effective approach to elucidate the physiological functions of expansin is comparison of morphological characteristics of control plants with their transgenic counterparts expressing sense and antisense constructs of target expansin genes (Cho and Cosgrove 2000). A number of previous studies showed a positive/negative effect of EXPA genes on leaf, petiole sizes and number of leaves (Cho and Cosgrove 2000; Choi et al. 2003; Gray-Mitsumune et al. 2008; Kuluev et al. 2012, 2013, 2014; Rochange et al. 2001). Hence, in this study, we validated the effect of overexpression of BrEXLB1 sense and antisense constructs driven by the CaMV35S promoter in transgenic Arabidopsis. Following hygromycin selection, eight independent T1 transgenic lines were randomly selected for leaf-morphology statistical analysis: four lines harboring the sense construct and four carrying the antisense construct, each differed in BrEXLB1 expression (Fig. 4b, c). Measurements were taken at three time points (15, 20, and 30 days after transferring the pots into the growth chamber). The blade width of the sense line pLSI9-7 was significantly higher than the wild type at day 30; whereas, the antisense line pLSI12-2 showed increased and decreased blade width at days 15 and 30, respectively, than the wild type and these values were statistically significant (Fig. 4e). Concurrently, the blade length of both sense and antisense lines at all three time points were not significantly different from that of wild type except antisense line pLSI12-4 which blade length was significantly lower than the wild type (Fig. 4d). Moreover, we observed inconsistent length in petioles of sense and antisense lines of BrEXLB1 transgenic Arabidopsis in comparison with wild types (Fig. 4f). The petiole length was significantly high in the sense lines of pLSI9-8 at day 20, pLSI9-7 at day 30 and in the antisense line of pLSI12-4 at day 15. Surprisingly, the sense line pLSI9-2 showed significantly decreased petiole length at day 30 than the wild type. mRNA accumulation not correlated with phenotypic changes observed in transgenic Arabidopsis. This is clear with sense line pLSI9-9 where relatively high number of BrEXLB1 transcripts found, but do not show any significant phenotypic change (Fig. 4). Moreover, the phenotypic changes induced by sense and antisense constructs are not uniform. However, it is evident that transgenic lines did affect the leaf morphology in comparison with wild types indicating other unknown factors might also be involved in leaf growth or morphology. The results also suggest that the ectopic expression of BrEXLB1 gene results differential and unstable pleiotropic effects on the growth of leaves. Interestingly, plant height were visually increased in BrEXLB1 (sense) overexpressing transgenic lines than wild type and BrEXLB1 antisense overexpressing lines. It is clear that BrEXLB1 either directly or indirectly regulate plant growth and development including leaf, and plant height/plant growth by its known cell wall loosening activity in Arabidopsis (Fig. 5). To understand the mechanisms behind BrEXLB1 mediated growth further in-depth analysis is necessary.
Moreover, the expression of GUS driven by promoter of BrEXLB1 in leaves and cotyledons of transgenic Arabidopsis was abundant while the expression of endogenous BrEXLB1 in B. rapa was undetectable. This difference in BrEXLB1 promoter activity suggests the likely interference of other factors and the different genetic constitution. Unlike in A. thaliana, three EXLB (EXLB1, 2 and 3) genes were present in B. rapa (Krishnamurthy et al.). Possibly, BrEXLB2 and BrEXLB3 would have direct or indirect effect in expression of BrEXLB1 in host plant, B. rapa. However, the effect of BrEXLB2 and BrEXLB3 is unlikely in transgenic Arabidopsis due to the absence of EXLB2 and EXLB3. In addition, other endogenous and exogenous factors including white light, red light, dark, IAA, JA and drought stress in B. rapa and A. thaliana would modulate the expression of BrEXLB1 differently as shown by transgenic promoter lines in the present study.
In conclusion, our findings suggest that BrEXLB1 may participate in germination, leaf development, and plant growth depending upon the phytohormone availability (esp. IAA, JA), interaction with other EXLB genes, transcriptional regulators, light types and intensity, dark periods and drought stress in a given physiological conditions. Moreover, BrEXLB1 may exhibit tissue preference as evidenced by GUS reporter-aided analysis of BrEXLB1-specific promoters in this study.
Abbreviations
- BrEXLB1 :
-
Brassica rapa expansin-like B1 gene
- GUS:
-
β-Glucuronidase
- IAA:
-
Indole acetic acid
- GA:
-
Gibberelic acid
- JA:
-
Jasmonic acid
- SA:
-
Salicylic acid
- ABA:
-
Abscisic acid
- qRT-PCR:
-
Quantitative reverse-transcription PCR
References
Abuqamar S, Ajeb S, Sham A et al (2013) A mutation in the expansin-like A2 gene enhances resistance to necrotrophic fungi and hypersensitivity to abiotic stress in Arabidopsis thaliana. Mol Plant Pathol 14:813–827. https://doi.org/10.1111/mpp.12049
Bae JM, Kwak MS, Noh SA et al (2014) Overexpression of sweetpotato expansin cDNA (IbEXP1) increases seed yield in Arabidopsis. Transgenic Res 23:657–667. https://doi.org/10.1007/s11248-014-9804-1
Bari R, Jones JDG (2009) Role of plant hormones in plant defence responses. Plant Mol Biol 69:473–488. https://doi.org/10.1007/s11103-008-9435-0
Brummell DA, Harpster MH, Dunsmuir P (1999) Differential expression of expansin genes family members during growth and ripening of tomato fruit. Plant Mol Biol 39:161–169. https://doi.org/10.1023/A:1006130018931
Chen Y, Han Y, Meng Z et al (2016) Overexpression of the wheat expansin gene TaEXPA2 improved seed production and drought tolerance in transgenic tobacco plants. PLoS ONE 11:1–24. https://doi.org/10.1371/journal.pone.0153494
Chen L, Zou W, Fei C et al (2018) α-Expansin EXPA4 positively regulates abiotic stress tolerance but negatively regulates pathogen resistance in Nicotiana tabacum. Plant Cell Physiol. https://doi.org/10.1093/pcp/pcy155
Cho HT (1997) Expression of expansin genes is correlated with growth in deepwater rice. Plant Cell Online 9:1661–1671. https://doi.org/10.1105/tpc.9.9.1661
Cho H-T, Cosgrove DJ (2000) Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc Natl Acad Sci 97:9783–9788. https://doi.org/10.1073/pnas.160276997
Choi D, Lee Y, Cho HT, Kende H (2003) Regulation of expansin gene expression affects growth and development in transgenic rice plants. Plant Cell 15:1386. https://doi.org/10.1105/tpc.011965.1998
Cosgrove DJ (2001) Wall structure and wall loosening. A look backwards and forwards. Plant Physiol 125:131–134. https://doi.org/10.1104/pp.125.1.131
Cosgrove DJ (2015) Plant expansins: diversity and interactions with plant cell walls. Curr Opin Plant Biol 25:162–172
Dal Santo S, Fasoli M, Cavallini E et al (2011) PhEXPA1, a Petunia hybrida expansin, is involved in cell wall metabolism and in plant architecture specification. Plant Signal Behav 6:2031–2034
Gray-Mitsumune M, Blomquist K, McQueen-Mason S et al (2008) Ectopic expression of a wood-abundant expansin PttEXPA1 promotes cell expansion in primary and secondary tissues in aspen. Plant Biotechnol J 6:62–72. https://doi.org/10.1111/j.1467-7652.2007.00295.x
Greenwood MS, Xu F, Hutchison KW (2006) The role of auxin-induced peaks of α-expansin expression during lateral root primordium formation in Pinus taeda. Physiol Plant 126:279–288. https://doi.org/10.1111/j.1399-3054.2006.00616.x
Harrison SJ, Mott EK, Parsley K et al (2006) A rapid and robust method of identifying transformed Arabidopsis thaliana seedlings following floral dip transformation. Plant Methods 2:1–7. https://doi.org/10.1186/1746-4811-2-19
Hong JK, Kim JS, Kim JA et al (2010) Identification and characterization of SHI family genes from Brassica rapa L. ssp. pekinensis. Genes Genomics 32:309–317. https://doi.org/10.1007/s13258-010-0011-z
Hong JK, Kim JA, Kim JS et al (2012) Overexpression of Brassica rapa SHI-RELATED SEQUENCE genes suppresses growth and development in Arabidopsis thaliana. Biotechnol Lett 34:1561–1569. https://doi.org/10.1007/s10529-012-0929-0
Hutchison KW, Singer PB, McInnis S et al (1999) Expansins are conserved in conifers and expressed in hypocotyls in response to exogenous auxin. Plant Physiol 120:827–832. https://doi.org/10.1104/pp.120.3.827
Kende H, Bradford KJ, Brummell DA et al (2004) Nomenclature for members of the expansin superfamily of genes and proteins. Plant Mol Biol 55:311–314. https://doi.org/10.1007/s11103-004-0158-6
Kim JH, Cho HT, Kende H (2000) Alpha-expansins in the semiaquatic ferns Marsilea quadrifolia and Regnellidium diphyllum: evolutionary aspects and physiological role in rachis elongation. Planta 212:85–92
Krishnamurthy P, Hong JK, Kim JA et al (2015) Genome-wide analysis of the expansin gene superfamily reveals Brassica rapa-specific evolutionary dynamics upon whole genome triplication. Mol Genet Genomics 290:521–530. https://doi.org/10.1007/s00438-014-0935-0
Kuluev BR, Knyazev AB, Lebedev YP, Chemeris AV (2012) Morphological and physiological characteristics of transgenic tobacco plants expressing expansin genes: AtEXP10 from Arabidopsis and PnEXPA1 from poplar. Russ J Plant Physiol 59:97–104. https://doi.org/10.1134/S1021443712010128
Kuluev BR, Safiullina MG, Knyazev AV, Chemeris AV (2013) Effect of ectopic expression of NtEXPA5 gene on cell size and growth of organs of transgenic tobacco plants. Russ J Dev Biol 44:28–34. https://doi.org/10.1134/S1062360413010049
Kuluev BR, Kniazev AV, Nikonorov IM, Chemeris AV (2014) Role of the expansin genes NtEXPA1 and NtEXPA4 in the regulation of cell extension during tobacco leaf growth. Russ J Genet 50:489–497
Lagerwerff JV, Ogata G, Eagle HE (1961) Control of osmotic pressure of culture solutions with polyethylene glycol. Science 133:1486–1487
Marowa P, Ding A, Kong Y (2016) Expansins: roles in plant growth and potential applications in crop improvement. Plant Cell Rep 35:949–965. https://doi.org/10.1007/s00299-016-1948-4
McQueen-Mason S, Cosgrove DJ (1994) Disruption of hydrogen bonding between plant cell wall polymers by proteins that induce wall extension. Proc Natl Acad Sci 91:6574–6578. https://doi.org/10.1073/pnas.91.14.6574
McQueen-Mason S, Durachko DM, Cosgrove DJ (1992) Two endogenous proteins that induce cell wall extension in plants. Plant Cell 4:1425. https://doi.org/10.2307/3869513
Perfus-Barbeoch L, Leonhardt N, Vavasseur A, Forestier C (2002) Heavy metal toxicity: cadmium permeates through calcium channels and disturbs the plant water status. Plant J 32:539–548. https://doi.org/10.1046/j.1365-313X.2002.01442.x
Rochange SF, Wenzel CL, Mcqueen-Mason SJ (2001) Impaired growth in transgenic plants over-expressing an expansin isoform. Plant Mol Biol 46:581–589. https://doi.org/10.1023/A:1010650217100
Sampedro J, Cosgrove DJ (2005) The expansin superfamily. Genome Biol 6:1–11. https://doi.org/10.1186/gb-2005-6-12-242
Santiago R, Pereira VM, De Souza WR et al (2018) Genome-wide identification, characterization and expression profile analysis of expansins gene family in sugarcane (Saccharum spp.). PLoS ONE 13:1–18. https://doi.org/10.1371/journal.pone.0191081
Sasidharan R, Voesenek LACJ, Pierik R (2011) Cell wall modifying proteins mediate plant acclimatization to biotic and abiotic stresses. Crit Rev Plant Sci 30:548–562. https://doi.org/10.1080/07352689.2011.615706
Wang T, Bum Y, Caporini MA et al (2013) Sensitivity-enhanced solid-state NMR detection of expansin’s target in plant cell walls. Proc Natl Acad Sci U S A 110:16444–16449
Wieczorek K, Golecki B, Gerdes L et al (2006) Expansins are involved in the formation of nematode-induced syncytia in roots of Arabidopsis thaliana. Plant J 48:98–112. https://doi.org/10.1111/j.1365-313X.2006.02856.x
Wrobel RL, Yoder JI (2001) Differential RNA expression of α-expansin gene family members in the parasitic angiosperm Triphysaria versicolor (Scrophulariaceae). Gene 266:85–93. https://doi.org/10.1016/S0378-1119(01)00376-6
Zhu Y, Wu N, Song W et al (2014) Soybean (Glycine max) expansin gene superfamily origins: segmental and tandem duplication events followed by divergent selection among subfamilies. BMC Plant Biol 14:1–19. https://doi.org/10.1186/1471-2229-14-93
Acknowledgements
This work was supported by the Rural Development Administration (Korea) through the Rural Program for Agricultural Science and Technology Development (Project No. PJ01247202) and the Next Generation Bio-green 21 Program (Project No. PJ01334002).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Krishnamurthy, P., Muthusamy, M., Kim, J.A. et al. Brassica rapa expansin-like B1 gene (BrEXLB1) regulate growth and development in transgenic Arabidopsis and elicits response to abiotic stresses. J. Plant Biochem. Biotechnol. 28, 437–446 (2019). https://doi.org/10.1007/s13562-019-00496-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13562-019-00496-1