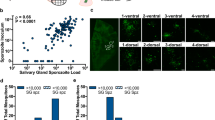
Abstract
The recent Zika virus (ZIKV) and chikungunya virus epidemics highlight the explosive nature of arthropod-borne viruses (arboviruses) transmitted by Aedes spp. mosquitoes1,2. Vector competence and the extrinsic incubation period (EIP) are two key entomological parameters used to assess the public health risk posed by arboviruses3. These are typically measured empirically by offering mosquitoes an infectious blood meal and temporally sampling mosquitoes to determine the infection and transmission status. This approach has been used for the better part of a century; however, it does not accurately capture the biology and behaviour of many mosquito vectors that refeed frequently (every 2–3 d)4. Here, we demonstrate that acquisition of a second non-infectious blood meal significantly shortens the EIP of ZIKV-infected Aedes aegypti by enhancing virus dissemination from the mosquito midgut. Similarly, a second blood meal increases the competence of this species for dengue virus and chikungunya virus as well as Aedes albopictus for ZIKV, suggesting that this phenomenon may be common among other virus–vector pairings and that A. albopictus might be a more important vector than once thought. Blood-meal-induced microperforations in the virus-impenetrable basal lamina that surrounds the midgut provide a mechanism for enhanced virus escape. Modelling of these findings reveals that a shortened EIP would result in a significant increase in the basic reproductive number, R0, estimated from experimental data. This helps to explain how A. aegypti can sustain explosive epidemics such as ZIKV despite relatively poor vector competence in single-feed laboratory trials. Together, these data demonstrate a direct and unrecognized link between mosquito feeding behaviour, EIP and vector competence.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Mayer, S. V., Tesh, R. B. & Vasilakis, N. The emergence of arthropod-borne viral diseases: a global prospective on dengue, chikungunya and Zika fevers. Acta Trop. 166, 155–163 (2017).
Faria, N. R. et al. Establishment and cryptic transmission of Zika virus in Brazil and the Americas. Nature 546, 406–410 (2017).
Kramer, L. D. & Ciota, A. T. Dissecting vectorial capacity for mosquito-borne viruses. Curr. Opin. Virol. 15, 112–118 (2015).
Scott, T. W. & Takken, W. Feeding strategies of anthropophilic mosquitoes result in increased risk of pathogen transmission. Trends Parasitol. 28, 114–121 (2012).
Weaver, S. C. & Reisen, W. K. Present and future arboviral threats. Antiviral Res. 85, 328–345 (2010).
Musso, D. & Gubler, D. J. Zika virus. Clin. Microbiol. Rev. 29, 487–524 (2016).
Scott, T. W. et al. Blood-feeding patterns of Aedes aegypti (Diptera: Culicidae) collected in a rural Thai village. J. Med. Entomol. 30, 922–927 (1993).
Scott, T. W. et al. Longitudinal studies of Aedes aegypti (Diptera: Culicidae) in Thailand and Puerto Rico: blood feeding frequency. J. Med. Entomol. 37, 89–101 (2000).
Ponlawat, A. & Harrington, L. C. Blood feeding patterns of Aedes aegypti and Aedes albopictus in Thailand. J. Med. Entomol. 42, 844–849 (2005).
Baak-Baak, C. M. et al. Blood feeding status, gonotrophic cycle and survivorship of Aedes (Stegomyia) aegypti (L.) (Diptera: Culicidae) caught in churches from Merida, Yucatan, Mexico. Neotrop. Entomol. 46, 622–630 (2017).
Sivan, A., Shriram, A. N., Sunish, I. P. & Vidhya, P. T. Host-feeding pattern of Aedes aegypti and Aedes albopictus (Diptera: Culicidae) in heterogeneous landscapes of South Andaman, Andaman and Nicobar Islands, India. Parasitol. Res. 114, 3539–3546 (2015).
De Benedictis, J. et al. Identification of the people from whom engorged Aedes aegypti took blood meals in Florida, Puerto Rico, using polymerase chain reaction-based DNA profiling. Am. J. Trop. Med. Hyg. 68, 437–446 (2003).
Barrera, R. et al. Vertebrate hosts of Aedes aegypti and Aedes mediovittatus (Diptera: Culicidae) in rural Puerto Rico. J. Med. Entomol. 49, 917–921 (2012).
Diagne, C. T. et al. Potential of selected Senegalese Aedes spp. mosquitoes (Diptera: Culicidae) to transmit Zika virus. BMC Infect. Dis. 15, 492 (2015).
Chouin-Carneiro, T. et al. Differential susceptibilities of Aedes aegypti and Aedes albopictus from the Americas to Zika virus. PLoS Negl. Trop. Dis. 10, e0004543 (2016).
Diagne, C. T. et al. Vector competence of Aedes aegypti and Aedes vittatus (Diptera: Culicidae) from Senegal and Cape Verde archipelago for West African lineages of chikungunya virus. Am. J. Trop. Med. Hyg. 91, 635–641 (2014).
Calvez, E. et al. Dengue-1 virus and vector competence of Aedes aegypti (Diptera: Culicidae) populations from New Caledonia. Parasites Vector 10, 381 (2017).
Diallo, M. et al. Vector competence of Aedes aegypti populations from Senegal for sylvatic and epidemic dengue 2 virus isolated in West Africa. Trans. R. Soc. Trop. Med. Hyg. 102, 493–498 (2008).
Black, W. Ct et al. Flavivirus susceptibility in Aedes aegypti. Arch. Med. Res. 33, 379–388 (2002).
Hardy, J. L. in The Arboviruses: Epidemiology and Ecology Vol. 1 (ed. Monath, T. P.) 87–126 (CRC, 1988).
Franz, A. W., Kantor, A. M., Passarelli, A. L. & Clem, R. J. Tissue barriers to arbovirus infection in mosquitoes. Viruses 7, 3741–3767 (2015).
Vazeille, M., Dehecq, J. S. & Failloux, A. B. Vectorial status of the Asian tiger mosquito Aedes albopictus of La Reunion Island for Zika virus. Med. Vet. Entomol. 32, 251–254 (2018).
O’Donnell, K. L., Bixby, M. A., Morin, K. J., Bradley, D. S. & Vaughan, J. A. Potential of a northern population of Aedes vexans (Diptera: Culicidae) to transmit Zika virus. J. Med. Entomol. 54, 1354–1359 (2017).
Okuda, K. et al. Cell death and regeneration in the midgut of the mosquito, Culex quinquefasciatus. J. Insect Physiol. 53, 1307–1315 (2007).
Dong, S. et al. Chikungunya virus dissemination from the midgut of Aedes aegypti is associated with temporal basal lamina degradation during bloodmeal digestion. PLoS Negl. Trop. Dis. 11, e0005976 (2017).
Kantor, A. M., Grant, D. G., Balaraman, V., White, T. A. & Franz, A. W. E. Ultrastructural analysis of chikungunya virus dissemination from the midgut of the yellow fever mosquito, Aedes aegypti. Viruses 10, 571 (2018).
Weaver, S. C., Scott, T. W., Lorenz, L. H., Lerdthusnee, K. & Romoser, W. S. Togavirus-associated pathologic changes in the midgut of a natural mosquito vector. J. Virol. 62, 2083–2090 (1988).
Delatte, H. et al. Blood-feeding behavior of Aedes albopictus, a vector of Chikungunya on La Reunion. Vector-Borne Zoonotic Dis. 10, 249–258 (2010).
Egizi, A., Healy, S. P. & Fonseca, D. M. Rapid blood meal scoring in anthropophilic Aedes albopictus and application of PCR blocking to avoid pseudogenes. Infect. Genet. Evol. 16C, 122–128 (2013).
Dickson, L. B., Sanchez-Vargas, I., Sylla, M., Fleming, K. & Black, W. C. 4th Vector competence in West African Aedes aegypti is flavivirus species and genotype dependent. PLoS Negl. Trop. Dis. 8, e3153 (2014).
Bosio, C. F., Beaty, B. J. & Black, W. C. 4th Quantitative genetics of vector competence for dengue-2 virus in Aedes aegypti. Am. J. Trop. Med. Hyg. 59, 965–970 (1998).
Kramer, L. D., Hardy, J. L., Presser, S. B. & Houk, E. J. Dissemination barriers for western equine encephalomyelitis virus in Culex tarsalis infected after ingestion of low viral doses. Am. J. Trop. Med. Hyg. 30, 190–197 (1981).
Houk, E. J., Hardy, J. L. & Chiles, R. E. Permeability of the midgut basal lamina in the mosquito, Culex tarsalis Coquillett (Insecta, Diptera). Acta Trop. 38, 163–171 (1981).
Perkins, T. A., Siraj, A. S., Ruktanonchai, C. W., Kraemer, M. U. G. & Tatem, A. J. Model-based projections of Zika virus infections in childbearing women in the Americas. Nat. Microbiol. 1, 16126.
Salazar, M. I., Richardson, J. H., Sanchez-Vargas, I., Olson, K. E. & Beaty, B. J. Dengue virus type 2: replication and tropisms in orally infected Aedes aegypti mosquitoes. BMC Microbiol. 7, 9 (2007).
Meyer, R. P., Hardy, J. L. & Presser, S. B. Comparative vector competence of Culex tarsalis and Culex quinquefasciatus from the Coachella, Imperial, and San Joaquin Valleys of California for St. Louis encephalitis virus. Am. J. Trop. Med. Hyg. 32, 305–311 (1983).
Turell, M. J. Reduced Rift Valley fever virus infection rates in mosquitoes associated with pledget feedings. Am. J. Trop. Med. Hyg. 39, 597–602 (1988).
Hsieh, P. & Robbins, P. W. Regulation of asparagine-linked oligosaccharide processing. Oligosaccharide processing in Aedes albopictus mosquito cells. J. Biol. Chem. 259, 2375–2382 (1984).
Armstrong, P. M. & Rico-Hesse, R. Differential susceptibility of Aedes aegypti to infection by the American and Southeast Asian genotypes of dengue type 2 virus. Vector-Borne Zoonotic Dis. 1, 159–168 (2001).
Airs, P. M., Kudrna, K. E. & Bartholomay, L. C. Impact of sugar composition on meal distribution, longevity, and insecticide toxicity in Aedes aegypti. Acta Trop. 191, 221–227 (2019).
Waggoner, J. J. et al. Viremia and clinical presentation in Nicaraguan patients infected with Zika virus, chikungunya virus, and dengue virus. Clin. Infect. Dis. 63, 1584–1590 (2016).
Musso, D. et al. Molecular detection of Zika virus in blood and RNA load determination during the French Polynesian outbreak. J. Med. Virol. 89, 1505–1510 (2017).
Haese, N. N. et al. Animal models of Chikungunya virus infection and disease. J. Infect. Dis. 214, S482–S487 (2016).
Anderson, J. F., Main, A. J., Delroux, K. & Fikrig, E. Extrinsic incubation periods for horizontal and vertical transmission of West Nile virus by Culex pipiens pipiens (Diptera: Culicidae). J. Med. Entomol. 45, 445–451 (2008).
Lanciotti, R. S. et al. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg. Infect. Dis. 14, 1232–1239 (2008).
Alm, E. et al. Universal single-probe RT-PCR assay for diagnosis of dengue virus infections. PLoS Negl. Trop. Dis. 8, e3416 (2014).
Lanciotti, R. S. et al. Chikungunya virus in US travelers returning from India, 2006. Emerg. Infect. Dis. 13, 764–767 (2007).
Chan, M. & Johansson, M. A. The incubation periods of Dengue viruses. PLoS ONE 7, e50972 (2012).
Ferguson, N. M. et al. Countering the Zika epidemic in Latin America. Science 353, 353–354 (2016).
Plummer, M. rjags: Bayesian Graphical Models using MCMC. R package version 4-8 (2018).
R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2014); http://www.R-project.org
Brooks, S. P. & Gelman, A. General methods for monitoring convergence of iterative simulations. J. Comput. Graph. Stat. 7, 434–455 (1998).
Christofferson, R. C. & Mores, C. N. Estimating the magnitude and direction of altered arbovirus transmission due to viral phenotype. PLoS ONE 6, e16298 (2011).
Smith, D. L. et al. Ross, Macdonald, and a theory for the dynamics and control of mosquito-transmitted pathogens. PLoS Pathog. 8, e1002588 (2012).
Chowell, G. et al. Using phenomenological models to characterize transmissibility and forecast patterns and final burden of Zika epidemics. PLoS Currents https://doi.org/10.1371/currents.outbreaks.f14b2217c902f453d9320a43a35b9583 (2016).
Funk, S. et al. Comparative analysis of dengue and Zika outbreaks reveals differences by setting and virus. PLoS Negl. Trop. Dis. 10, e0005173 (2016).
Carnell, R. lhs: Latin Hypercube Samples. R package version 1.0.1 (2019).
Acknowledgements
We thank M. C. Thomas, M. Olson, T. Petruff and M. Correa who provided technical assistance with rearing, processing and virus testing of mosquitoes. This publication was supported by the Cooperative Agreement Number U01CK000509, funded by the Centers for Disease Control and Prevention. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the Centers for Disease Control and Prevention or the Department of Health and Human Services. This work was further supported by the US Department of Agriculture Hatch Funds and Multistate Research Project (CONH00773 and NE1443), the National Center for Advancing Translational Sciences of the National Institutes of Health (CTSA grant number UL1 TR001863 and KL2 TR001862) (J.L.W.) and the National Institutes of Health/National Institute of Allergy and Infectious Diseases (R01 AI112970) (V.E.P.) and R21 AI129464 (B.D.F.).
Author information
Authors and Affiliations
Contributions
P.M.A., H.Y.E., V.E.P. and D.E.B. conceived and designed the experiments and wrote the manuscript. P.M.A., D.E.B., T.M., M.R.M., B.D.F., A.B., M.J.M. and A.G.-S. performed mosquito experiments including mosquito infections, dissections, transmission assays and the quantification of viral RNA and infectious virus particles. D.E.B. performed the retrograde infection assay and fluorescence microscopy. P.J.C. performed the CHP assay. J.J.S. and T.G.A. performed the scanning electron microscopy. P.M.A. and D.E.B. statistically analysed and interpreted the experimental data. H.Y.E., J.L.W. and V.E.P. performed the modelling and statistics associated with the modelling. P.M.A., V.E.P., B.D.F. and D.E.B. oversaw the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Non-infectious bloodmeals prior to infectious bloodmeals do not increase dissemination rates.
(a) Schematic of experimental design. At (b) 4, (c) 6, and (d) 9 dpi, midguts and legs were dissected from both cohorts and tested for the presence of ZIKV RNA by RT-qPCR. (●) single feed; (●) double feed. Data were analyzed by Two-sided Fisher’s exact test. Sample sizes (represented as a fraction of positive samples/ total samples) for each treatment/ timepoint are embedded in the figures above each experimental group. Center values represent the proportion and error bars represent the binomial SE of sample proportions.
Extended Data Fig. 2 Increased transmission of ZIKV RNA and infectious virions associated with acquisition of an additional non-infectious bloodmeal.
Aedes aegypti mosquitoes were offered a ZIKV infectious bloodmeal and at 3 dpi individuals in the double-feed groups were fed a second, non-infectious bloodmeal. 10 dpi mosquito saliva was collected and assayed for ZIKV RNA and infectious virions by RT-qPCR and plaque assay respectively. (●) single feed; (●) double feed. Data were analyzed by Two-sided Fisher’s exact test. (*) p<0.05. Sample sizes (represented as a fraction of positive samples/ total samples) for each treatment/ timepoint are embedded in the figures above each experimental group. Center values represent the proportion and error bars represent the binomial SE of sample proportions.
Extended Data Fig. 3 Multiple feeding events increase the potential of Aedes albopictus to transmit ZIKV.
(a) Schematic of experimental design. Paired bodies (MGI; midgut infection; % of mosquitoes with viral RNA in their bodies), legs (DI; disseminated infection; % of body positive mosquitoes with viral RNA in their legs) and salivary glands (SGI; salivary gland infection; % of leg positive mosquitoes with viral RNA in their salivary glands) were collected and assayed for the presence of viral RNA (b) 7 dpi and (c) 10 dpi. (d) SGI data from (b) and (c) analyzed as the % of ZIKV-exposed mosquitoes with a salivary gland infection. The data presented represents at least three experimental replicates. (●) single-feed; (●) double-feed. Data were analyzed by Two-sided Fisher’s exact test. (*) p<0.05, (**) p<0.01, (***) p<0.001. Sample sizes (represented as a fraction of positive samples/ total samples) for each treatment/ timepoint are embedded in the figures above each experimental group. Center values represent the proportion and error bars represent the binomial SE of sample proportions.
Extended Data Fig. 4 Increased dissemination rates associated with multiple feeding are not due to increased midgut replication.
Aedes aegypti mosquitoes were offered a ZIKV-infectious bloodmeal and 4 dpi individuals in the double feed groups were fed a second, non-infectious bloodmeal. At 7 dpi, mosquito midguts and legs were dissected and viral genomic equivalents were determined. (a) Midgut genome equivalents based on feeding status regardless if the infection was restricted to the midgut or disseminated (n=10/ group). (b) Midgut genome equivalents based on infection status (disseminated (n=14) vs. not disseminated (n=6) (that is midgut restricted)) regardless of feeding status. (●) single feed; (●) double feed. Data were analyzed by Two-sided T-test. Center values represent means and error bars represent SD.
Extended Data Fig. 5 Distributions of mean EIP estimates.
(●) experimental single-feed; (●) meta-analysis single-feed; (●) double feed, from a thinned subset (10,000 iterations) of each model’s respective posterior shape and rate estimates.
Extended Data Fig. 6 Comparison of R0 estimates from experimental data to published estimates from field studies.
Circles correspond to point estimates of R0 derived from an individual study and setting and/or methodology, while the lines show the corresponding 95% CI (n=41 estimates from 22 studies). Colors are used to show estimates from different regions (light blue: Oceania, dark blue: South America, dark green: Central America, gray: other) and our own experimental results (blue: single-feed model, red: double-feed model).
Extended Data Fig. 7 Results of a best/worst case scenario sensitivity analysis and a one-way random sampling sensitivity analysis.
(a) Scenario sensitivity analysis assessing the effect of varying each parameter according to its lowest and highest bounds on the difference in \(R_0\left( {{\it{R}}_{0_{DF}} - {\it{R}}_{0_{SF}}} \right)\) (blue=lowest value; red=highest value for each parameter). The initial difference of 0.71 was obtained by holding all parameters constant at their mean or given value (Supplementary Table 4). (b) Violin plots show the probably density of the difference in R0 \(\left( {{\it{R}}_{0_{DF}} - {\it{R}}_{0_{SF}}} \right)\) when we randomly sampled each parameter 10,000 times from its specified distribution (Supplementary Table 4), while holding all other parameters constant. Each violin spans the 98% quantile of the distribution of R0 differences with the width proportional to the probability of observing a particular value of difference. The horizontal line at 0.71 represents the difference in R0 when all parameters are held constant at their mean or given value.
Extended Data Fig. 8 Comparison of R0 and p.
(a) The mean value for 10,000 simulations of R0 is plotted for every value of p (blue= single-feed model, red= double-feed model, dashed black= reference line at R0=1), along with (b) the proportion of those simulations for which R0 >1.
Extended Data Fig. 9 Salivary Gland Infection Meta-Analysis.
Salivary Gland Infection (SGI) data aggregated from 8 published studies are plotted, with each observation (n=45) weighted by sample size and color-coded by study. The black line shows the best-fit gamma CDF model for salivary gland data and the blue line for combined meta-analysis single-feed dissemination data (data points not shown).
Extended Data Fig. 10 Results of an uncertainty analysis assessing the distribution of R0 under different feeding assumptions.
(a) We compared histograms of the single-feed R0 distribution specified by the uncertainty analysis (blue) and double-feed R0 distribution (red) and the overlap (purple) generated from 10,000 iterations of Latin Hypercube Sampling. The mean R0 for each respective distribution is also shown as a horizontal line (\({\it{R}}_{0_{SF}} = 6.68\), \({\it{R}}_{0_{DF}} = 7.55\)). (b) The distribution of the difference in \(R_0\left( {{\it{R}}_{0_{DF}} - {\it{R}}_{0_{SF}}} \right)\) is plotted for each random sample (n=10,000).
Supplementary information
Supplementary Information
Supplementary Tables 1–6 and Supplementary References.
Source data
Source Data Fig. 1
Replicate data for Figure 1 with sample sizes and precise P values.
Source Data Fig. 2
Raw data for figure 2c with sample sizes and precise P values.
Rights and permissions
About this article
Cite this article
Armstrong, P.M., Ehrlich, H.Y., Magalhaes, T. et al. Successive blood meals enhance virus dissemination within mosquitoes and increase transmission potential. Nat Microbiol 5, 239–247 (2020). https://doi.org/10.1038/s41564-019-0619-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-019-0619-y
This article is cited by
-
Co-infection of dengue and Zika viruses mutually enhances viral replication in the mosquito Aedes aegypti
Parasites & Vectors (2023)
-
Ad libitum consumption of protein- or peptide-sucrose solutions stimulates egg formation by prolonging the vitellogenic phase of oogenesis in anautogenous mosquitoes
Parasites & Vectors (2022)
-
The microbiome and mosquito vectorial capacity: rich potential for discovery and translation
Microbiome (2021)
-
Population bottlenecks and founder effects: implications for mosquito-borne arboviral emergence
Nature Reviews Microbiology (2021)
-
Mapping the cryptic spread of the 2015–2016 global Zika virus epidemic
BMC Medicine (2020)