Abstract
Recognition of histone-modified nucleosomes by specific reader domains underlies the regulation of chromatin-associated processes. Whereas structural studies revealed how reader domains bind modified histone peptides, it is unclear how reader domains interact with modified nucleosomes. Here, we report the cryo-electron microscopy structure of the PWWP reader domain of human transcriptional coactivator LEDGF in complex with an H3K36-methylated nucleosome at 3.2–Å resolution. The structure reveals multivalent binding of the reader domain to the methylated histone tail and to both gyres of nucleosomal DNA, explaining the known cooperative interactions. The observed cross-gyre binding may contribute to nucleosome integrity during transcription. The structure also explains how human PWWP domain-containing proteins are recruited to H3K36-methylated regions of the genome for transcription, histone acetylation and methylation, and for DNA methylation and repair.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Patel, D. J. & Wang, Z. Readout of epigenetic modifications. Annu. Rev. Biochem. 82, 81–118 (2013).
Weaver, T., Morrison, E. & Musselman, C. Reading more than histones: the prevalence of nucleic acid binding among reader domains. Molecules 23, 2614 (2018).
Marabelli, C. et al. A tail-based mechanism drives nucleosome demethylation by the LSD2/NPAC multimeric complex. Cell Rep. 27, 387–399.e7 (2019).
Stec, I. et al. WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf-Hirschhorn syndrome critical region and is fused to IgH in t(1;14) multiple myeloma. Hum. Mol. Genet. 7, 1071–1082 (1998).
Izumoto, Y., Kuroda, T., Harada, H., Kishimoto, T. & Nakamura, H. Hepatoma-derived growth factor belongs to a gene family in mice showing significant homology in the amino terminus. Biochem. Biophys. Res. Commun. 238, 26–32 (1997).
Stec, I., Nagl, S. B., van Ommen, G. J. B. & den Dunnen, J. T. The PWWP domain: a potential protein-protein interaction domain in nuclear proteins influencing differentiation? FEBS Lett. 473, 1–5 (2000).
Qiu, C., Sawada, K., Zhang, X. & Cheng, X. D. The PWWP domain of mammalian DNA methyltransferase Dnmt3b defines a new family of DNA-binding folds. Nat. Struct. Biol. 9, 217–224 (2002).
Rona, G. B., Eleutherio, E. C. & Pinheiro, A. S. PWWP domains and their modes of sensing DNA and histone methylated lysines. Biophys. Rev. 8, 63–74 (2016).
Hughes, R. M., Wiggins, K. R., Khorasanizadeh, S. & Waters, M. L. Recognition of trimethyllysine by a chromodomain is not driven by the hydrophobic effect. Proc. Natl Acad. Sci. USA 104, 11184–11188 (2007).
Maurer-Stroh, S. et al. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem. Sci. 28, 69–74 (2003).
Vezzoli, A. et al. Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nat. Struct. Mol. Biol. 17, 617–619 (2010).
Dhayalan, A. et al. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J. Biol. Chem. 285, 26114–26120 (2010).
Vermeulen, M. et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 142, 967–980 (2010).
Pradeepa, M. M., Sutherland, H. G., Ule, J., Grimes, G. R. & Bickmore, W. A. Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. PLoS Genet. 8, e1002717 (2012).
Wu, H. et al. Structural and histone binding ability characterizations of human PWWP domains. PLoS ONE 6, e18919 (2011).
Sankaran, S. M., Wilkinson, A. W., Elias, J. E. & Gozani, O. A PWWP domain of histone-lysine N-methyltransferase NSD2 binds to dimethylated Lys-36 of histone H3 and regulates NSD2 function at chromatin. J. Biol. Chem. 291, 8465–8474 (2016).
Barski, A. et al. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (2007).
Li, J., Moazed, D. & Gygi, S. P. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J. Biol. Chem. 277, 49383–49388 (2002).
Sun, X.-J. et al. Identification and characterization of a novel human histone H3 lysine 36-specific methyltransferase. J. Biol. Chem. 280, 35261–35271 (2005).
Wagner, E. J. & Carpenter, P. B. Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell Biol. 13, 115–126 (2012).
Li, H. et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 442, 91–95 (2006).
Ge, Y. Z. et al. Chromatin targeting of de novo DNA methyltransferases by the PWWP domain. J. Biol. Chem. 279, 25447–25454 (2004).
Qiu, Y. et al. Solution structure of the Pdp1 PWWP domain reveals its unique binding sites for methylated H4K20 and DNA. Biochem. J. 442, 527–538 (2012).
Eidahl, J. O. et al. Structural basis for high-affinity binding of LEDGF PWWP to mononucleosomes. Nucleic Acids Res. 41, 3924–3936 (2013).
van Nuland, R. et al. Nucleosomal DNA binding drives the recognition of H3K36-methylated nucleosomes by the PSIP1-PWWP domain. Epigenetics Chromatin 6, 12 (2013).
Tian, W. et al. The HRP3 PWWP domain recognizes the minor groove of double-stranded DNA and recruits HRP3 to chromatin. Nucleic Acids Res. 47, 5436–5448 (2019).
Turlure, F., Maertens, G., Rahman, S., Cherepanov, P. & Engelman, A. A tripartite DNA-binding element, comprised of the nuclear localization signal and two AT-hook motifs, mediates the association of LEDGF/p75 with chromatin in vivo. Nucleic Acids Res. 34, 1653–1665 (2006).
Llano, M. et al. Identification and characterization of the chromatin-binding domains of the HIV-1 integrase interactor LEDGF/p75. J. Mol. Biol. 360, 760–773 (2006).
Ge, H., Si, Y. Z. & Roeder, R. G. Isolation of cDNAs encoding novel transcription coactivators p52 and p75 reveals an alternate regulatory mechanism of transcriptional activation. EMBO J. 17, 6723–6729 (1998).
LeRoy, G. et al. LEDGF and HDGF2 relieve the nucleosome-induced barrier to transcription. Sci. Adv. 5, eaay3068 (2019).
Simon, M. D. et al. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell 128, 1003–1012 (2007).
Tsutsui, K. M., Sano, K., Hosoya, O., Miyamoto, T. & Tsutsui, K. Nuclear protein LEDGF/p75 recognizes supercoiled DNA by a novel DNA-binding domain. Nucleic Acids Res. 39, 5067–5081 (2011).
Hendrix, J. et al. The transcriptional co-activator LEDGF/p75 displays a dynamic scan-and-lock mechanism for chromatin tethering. Nucleic Acids Res. 39, 1310–1325 (2011).
Makde, R. D., England, J. R., Yennawar, H. P. & Tan, S. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature 467, 562–566 (2010).
Wen, H. et al. ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature 508, 263–268 (2014).
Rondelet, G., Dal Maso, T., Willems, L. & Wouters, J. Structural basis for recognition of histone H3K36me3 nucleosome by human de novo DNA methyltransferases 3A and 3B. J. Struct. Biol. 194, 357–367 (2016).
Qin, S. & Min, J. R. Structure and function of the nucleosome-binding PWWP domain. Trends Biochem. Sci. 39, 536–547 (2014).
McGinty, R. K. & Tan, S. Recognition of the nucleosome by chromatin factors and enzymes. Curr. Opin. Struct. Biol. 37, 54–61 (2016).
Morgan, M. T. et al. Structural basis for histone H2B deubiquitination by the SAGA DUB module. Science 351, 725–728 (2016).
Worden, E. J., Hoffmann, N. A., Hicks, C. W. & Wolberger, C. Mechanism of cross-talk between H2B ubiquitination and H3 methylation by Dot1L. Cell 176, 1490–1501.e12 (2019).
Shun, M.-C. et al. Identification and characterization of PWWP domain residues critical for LEDGF/p75 chromatin binding and human immunodeficiency virus type 1 infectivity. J. Virol. 82, 11555–11567 (2008).
Baubec, T. et al. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature 520, 243–247 (2015).
Li, F. et al. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSα. Cell 153, 590–600 (2013).
Wolffe, A. P. Architectural transcription factors. Science 264, 1100–1101 (1994).
Zhu, F. et al. The interaction landscape between transcription factors and the nucleosome. Nature 562, 76–81 (2018).
Maskell, D. P. et al. Structural basis for retroviral integration into nucleosomes. Nature 523, 366–369 (2015).
Farnung, L., Vos, S. M., Wigge, C. & Cramer, P. Nucleosome–Chd1 structure and implications for chromatin remodelling. Nature 550, 539–542 (2017).
Eustermann, S. et al. Structural basis for ATP-dependent chromatin remodelling by the INO80 complex. Nature 556, 386–390 (2018).
Ayala, R. et al. Structure and regulation of the human INO80–nucleosome complex. Nature 556, 391–395 (2018).
Willhoft, O. et al. Structure and dynamics of the yeast SWR1-nucleosome complex. Science 362, eaat7716 (2018).
Venkatesh, S. et al. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature 489, 452–455 (2012).
Venkatesh, S. & Workman, J. L. Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 16, 178–189 (2015).
Carrozza, M. J. et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123, 581–592 (2005).
Smolle, M. et al. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nat. Struct. Mol. Biol. 19, 884–892 (2012).
Zhang, P. et al. Structure of human MRG15 chromo domain and its binding to Lys36-methylated histone H3. Nucleic Acids Res. 34, 6621–6628 (2006).
Musselman, C. A. et al. Molecular basis for H3K36me3 recognition by the Tudor domain of PHF1. Nat. Struct. Mol. Biol. 19, 1266–1272 (2012).
Ballare, C. et al. Phf19 links methylated Lys36 of histone H3 to regulation of Polycomb activity. Nat. Struct. Mol. Biol. 19, 1257–1265 (2012).
Brien, G. L. et al. Polycomb PHF19 binds H3K36me3 and recruits PRC2 and demethylase NO66 to embryonic stem cell genes during differentiation. Nat. Struct. Mol. Biol. 19, 1273–1281 (2012).
Musselman, C. A. et al. Binding of PHF1 Tudor to H3K36me3 enhances nucleosome accessibility. Nat. Commun. 4, 2969 (2013).
Kim, D. et al. Corecognition of DNA and a methylated histone tail by the MSL3 chromodomain. Nat. Struct. Mol. Biol. 17, 1027–1029 (2010).
Connelly, K. E. et al. Engagement of DNA and H3K27me3 by the CBX8 chromodomain drives chromatin association. Nucleic Acids Res. 47, 2289–2305 (2019).
Jacobson, R. H., Ladurner, A. G., King, D. S. & Tjian, R. Structure and function of a human TAFII250 double bromodomain module. Science 288, 1422–1425 (2000).
Miller, T. C. et al. A bromodomain-DNA interaction facilitates acetylation-dependent bivalent nucleosome recognition by the BET protein BRDT. Nat. Commun. 7, 13855 (2016).
Morrison, E. A. et al. DNA binding drives the association of BRG1/hBRM bromodomains with nucleosomes. Nat. Commun. 8, 16080 (2017).
Dyer, P. N. et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 375, 23–44 (2004).
Tegunov, D. & Cramer, P. Real-time cryo-EM data pre-processing with Warp. Nat. Methods 16, 1146–1152 (2019).
Zivanov, J. et al. RELION-3: new tools for automated high-resolution cryo-EM structure determination. eLife 7, e42166 (2018).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290 (2017).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Acknowledgements
We thank C. Oberthür for help with LEDGF protein purification, U. Neef and S. Vos for maintaining insect cell stocks and S. Aibara for helpful discussion. H.W. was supported by an EMBO Long-Term Fellowship (grant no. ALTF 650-2017). P.C. was supported by the Deutsche Forschungsgemeinschaft (grant nos. SFB860, SPP1935, SPP2191), the Advanced Grant ‘TRANSREGULON’ from the European Research Council (grant agreement no. 693023) and the Volkswagen Foundation.
Author information
Authors and Affiliations
Contributions
H.W. and L.F. initiated the project. H.W. designed and conducted all of the experiments and data analysis unless stated otherwise. L.F. cloned and purified full-length LEDGF protein. C.D. maintained the EM facility and advised on microscope setup. P.C. supervised research. H.W. and P.C. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Anke Sparmann was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
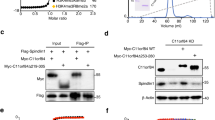
Extended Data Fig. 1 Binding of LEDGF to H3KC36me3-modified nucleosome.
a, EMSA reveals that full-length LEDGF preferentially binds to a H3KC36me3-modified nucleosome with longer (165 bp) DNA. Molar ratio of full-length LEDGF to indicated nucleosomes are shown on the top of each lane. Bands correspond to each component and complexes are labeled on the right. Bands of degraded LEDGF-bound nucleosomes are denoted with *. For gel source data, see Source Data Extended Data Fig. 1. b, Reconstructed EM density maps of 145bp and 165bp H3KC36me3-modified nucleosome with LEDGF. Note that the presence of the extra DNA in the latter complex leads to a defined additional density for the PWWP domain. c, Mass spectrometry measurement of the H3KC36me3 modified histone H3. Left: molecular weight measurement of H3K36C mutant. Right: molecular weight measurement of H3KC36me3 modified H3.
Extended Data Fig. 2 Cryo-EM data processing.
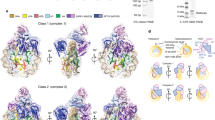
a, Data processing procedure for the complex of the 165 bp H3KC36me3-modified nucleosome with LEDGF using Warp and Relion. b, Fourier Shell Correlation (FSC) plot for the reconstruction using 55,142 particles in the indicated class (enclosed by dashed line in the final step in a). The overall resolution is 3.2 Å as determined by the FSC 0.143 criterion. c, Local resolution assessment of the final cryo-EM map. d, Euler angle distribution of particles used in the final 3D reconstruction.
Extended Data Fig. 3 Cryo-EM density.
a, A vertical slice through the structure. Models of all chains are shown as sticks and the cryo-EM density is shown as a gray mesh. b, A horizontal slice through the structure. Models of all chains are shown as sticks and the EM density is shown as a gray mesh. c, Density of histone H3 residue KC36me3 and its interacting residues of the aromatic cage in the PWWP domain. d, Density of DNA-interacting residues of patch 1. e, Density of part of the nucleosomal DNA at SHL 0. f, Density of the dyad DNA base pair. g, Density of the B-factor sharpened (left) and unsharpened (right) PWWP domain.
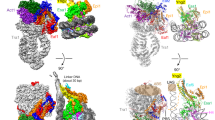
Extended Data Fig. 4 Nucleosome-PWWP interface and comparison with other PWWP-DNA structures.
a, Nucleosome-PWWP interface. Residues colored in white recognize H3KC36me3 and residues colored in green interact with DNA. b, Front and side view of DNA conformation comparison with other known PWWP-DNA structures. PDB code of the structures used are: 5XSK (HDGF) and 6IIS (HDGF3L, also known as HRP3). c, Schematic view of DNA interactions. Electrostatic interactions and hydrogen bonds are shown as yellow dashes. SHLs are denoted.
Extended Data Fig. 5 Comparison of the location of the PWWP domain in our nucleosome-PWWP complex structure with previously proposed models.
a, Front view of the comparison with two models proposed for LEDGF (gray) 25 or its highly conserved homolog HDGFL3 (yellow)26 with our structure (pink). Whereas in one model (yellow) the domain is rotated by around 180 degrees and shifted to SHL −1 on one DNA gyre, in another model (Gray) the domain is moved to SHL +6.5 and −1.5, and placed in the major groove of the DNA gyres. b, Side view of the comparison shown in panel a.
Extended Data Fig. 6 Comparison with other ‘royal’ family domains bound to methylated H3K36 peptides.
a, Structures of the PWWP, Tudor and chromo domain bound with methylated H3K36 peptides. PDB codes of structures used here are: 4HCZ (PHF1), 2F5K (MRG15) and 4PLI (H3K36me3 of MRG2). b, Superposition of all three structures shown in a. c, Placement of the PHF1 Tudor domain structure (yellow)60 onto our nucleosome-PWWP structure based on superposition of the H3 peptides in both structures reveals a clash between the Tudor domain and the nucleosomal DNA (red dashed circle). This shows that the Tudor domain must bind differently, and may unwind the end of nucleosomal DNA or alter the conformation of the H3 tail, or both.
Supplementary information
Supplementary Information
Supplementary Note of sequence alignment.
Source data
Source Data Extended Data Fig. 1
Uncropped gel.
Rights and permissions
About this article
Cite this article
Wang, H., Farnung, L., Dienemann, C. et al. Structure of H3K36-methylated nucleosome–PWWP complex reveals multivalent cross-gyre binding. Nat Struct Mol Biol 27, 8–13 (2020). https://doi.org/10.1038/s41594-019-0345-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-019-0345-4
This article is cited by
-
DNMT3B PWWP mutations cause hypermethylation of heterochromatin
EMBO Reports (2024)
-
Structure of histone deacetylase complex Rpd3S bound to nucleosome
Nature Structural & Molecular Biology (2023)
-
Structures of transcription preinitiation complex engaged with the +1 nucleosome
Nature Structural & Molecular Biology (2023)
-
Structure of the complete Saccharomyces cerevisiae Rpd3S-nucleosome complex
Nature Communications (2023)
-
Base editor scanning charts the DNMT3A activity landscape
Nature Chemical Biology (2023)