Abstract
Introduction
This paper presents a lifelong learning framework which constantly adapts with changing data patterns over time through incremental learning approach. In many big data systems, iterative re-training high dimensional data from scratch is computationally infeasible since constant data stream ingestion on top of a historical data pool increases the training time exponentially. Therefore, the need arises on how to retain past learning and fast update the model incrementally based on the new data. Also, the current machine learning approaches do the model prediction without providing a comprehensive root cause analysis. To resolve these limitations, our framework lays foundations on an ensemble process between stream data with historical batch data for an incremental lifelong learning (LML) model.
Case description
A cancer patient’s pathological tests like blood, DNA, urine or tissue analysis provide a unique signature based on the DNA combinations. Our analysis allows personalized and targeted medications and achieves a therapeutic response. Model is evaluated through data from The National Cancer Institute’s Genomic Data Commons unified data repository. The aim is to prescribe personalized medicine based on the thousands of genotype and phenotype parameters for each patient.
Discussion and evaluation
The model uses a dimension reduction method to reduce training time at an online sliding window setting. We identify the Gleason score as a determining factor for cancer possibility and substantiate our claim through Lilliefors and Kolmogorov–Smirnov test. We present clustering and Random Decision Forest results. The model’s prediction accuracy is compared with standard machine learning algorithms for numeric and categorical fields.
Conclusion
We propose an ensemble framework of stream and batch data for incremental lifelong learning. The framework successively applies first streaming clustering technique and then Random Decision Forest Regressor/Classifier to isolate anomalous patient data and provides reasoning through root cause analysis by feature correlations with an aim to improve the overall survival rate. While the stream clustering technique creates groups of patient profiles, RDF further drills down into each group for comparison and reasoning for useful actionable insights. The proposed MALA architecture retains the past learned knowledge and transfer to future learning and iteratively becomes more knowledgeable over time.
Similar content being viewed by others
Introduction
Motivation
Using the past acquired knowledge to improve future learning leads to the next generation of machine learning. Lifelong learning mimics incremental and always learning experience of human life. Training a large volume of a historical dataset requires large cluster and expensive infrastructure set up to make the long-running one-shot batch jobs unrealistic on a big data pile. Batch tasks are not agile enough to adapt quickly with each new wave of ever arriving datasets. Hadoop and its de facto MapReduce framework make the learning process wait until a full set of data is collected and hours or even days long batch jobs get completed. On the other hand, streaming learning enables models to get trained and updated after each passing mini-batch of a window. So, the model can catch up to the newest trends faster and with a much smaller cluster size, reducing the infrastructure overhead significantly. But streaming learning gets overwhelmed with a large set of the historical data pool, to begin with. To address the responsiveness issue to the traditional batch model, in this paper, we introduce a mix-model approach through a big data Lambda Architecture that considers batch and stream as two collaborating multi-agent systems. To start with, the batch system saves its learning to the Hadoop Distributed File System (HDFS) for later use by stream engine. The streaming system can then update the model incrementally. The streaming model initializes itself with saved learning from the batch by loading the trained model from HDFS into Apache Spark Discretized Stream (DStream). Through a configurable mini-batch of the time window of few hours, the model gets re-trained and at the same time, predicts continually on test data. The updated model through stream engine is persisted and replicated into HDFS at a periodic interval of 6 h to run any ad-hoc batch query. Additional static data can be merged to stored HDFS data by a stream processor and the mini-batch can pick and continue from thereon.
Lambda Architecture
Lambda Architecture (LA) [1] is a simultaneous hybrid processing approach, enabling access to low latency real-time frameworks along with a high throughput MapReduce batch framework over a distributed setup. New data is fed into both the batch and stream layers simultaneously with an objective to serve both real-time events with low-latency responses while a comprehensive analysis of the data is done through the batch pipeline. See Fig. 1.
The Lambda Architecture combines low latency real-time frameworks with a comprehensive, high throughput MapReduce batch framework through distributed frameworks like Hadoop and Spark. Observe, while data is processed real-time as Spark DStreams, simultaneous batch processing takes place through the stored data from HDFS. A NoSQL store (Cassandra) stores the consolidated view from stream and batch
Our contributions
Through the period of the last two decades, there has been significant progress in machine learning algorithms and frameworks. However, much less emphasis was put on how these methods and algorithms can be used to train over an extended period of time to incrementally become more knowledgeable through knowledge retainment and transfer. Also, the current predictive analysis frameworks provide forecasting without much explanation and root cause analysis. We addressed these research gaps between just analytics and more explicit actionable insights through reasoning and a truly lifelong learning platform. The framework is effective in high dimensional big data applications through its Apache spark in-memory data processing capability, unique dimension reduction technique and incremental never-ending learning approach as outlined below:
Lifelong learning model
In this work, we propose a Multi-agent Lambda Architecture (MALA), a collaborative ensemble framework for stream and batch data. Initializing with batch processing results as offset, the framework successively applies (i) a streaming clustering procedure to group the target data points, followed by (ii) a Random Decision Forest Regressor and Classification algorithm to provide reasoning through dynamic root cause analysis, compare between groups and performs forecasting. The model’s real-time and batch agents use Spark MlLib APIs. In the MALA framework, a Knowledge Miner (KM) consolidates the past learning with recent stream updates into a Knowledge Base (KB), making the model a Lifelong Learning System.
Streaming clustering through dimension reduction
The objective is to achieve clusters of similar data profiles and isolate the specific clusters for further investigations. The basic idea for an online version of K-Means clustering is to divide the data stream into mini-batch windows and to incorporate knowledge learned in the previous window into the following ones. Hence, in streaming K-means clustering, the model is updated with each rolling window based on a combination between cluster centers computed from the preceding mini-batches and the current mini-batch. The framework further enables the merging of a large static historical data pool with the latest and most updated streaming model. We adapt to a unique dimension reduction method of the feature vector to drastically reduce the model training time.
Random Decision Forest-based forecasting and root cause analysis
A Random Decision Forest Regressor graph further drills down into each anomalous cluster and provides a comprehensive root cause analysis and prediction for future items. The proposed framework produces the Decision Tree graph in JSON formatted data as well as in a tree structure. A number of optimizations are proposed for the Random Decision Forest model including data sampling, important feature selection, and optimal Decision Tree hyperparameters to improve the training time through reduced data volume without affecting the overall accuracy.
We present a unique dimension reduction method of feature vectors enabling quick re-training for clustering and Random Decision Forest through the MALA architecture. With this approach, the batch model creates the offset (or initial point). On each streaming window of the dataset, the streaming model just needs to learn the updated features or corrections during the re-training process.
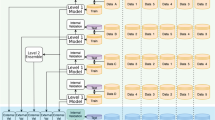
The two-stage process of the proposed architecture is depicted in Fig. 2.
The End-to-end flow of the proposed architecture. The method proceeds in two stages. (i) A hybrid clustering procedure creates a similar data profile and detects outliers. (ii) A Random Decision Forest process creates a decision graph on each anomalous clusters providing reasoning with root cause analysis and forecasting for future items
Related work
Our work addresses a problem that is analogous to and has gathered attention from several machine learning models such as lifelong learning, incremental learning, multi-task learning, transfer learning, and streaming learning. Thrun [1] first studied supervised lifelong learning through the decade of the 90s. The work explored the information sharing across multiple collaborating tasks through neural network binary classification. A neural network approach to LML was introduced and subsequently improved by Silver et al. through the years 1996 to 2015 [2,3,4,5,6,7]. Cumulative learning is explored in the form of LML which builds a classifier to classify all the previous and the new classes by updating the old classifier [8]. Ruvolo et al. [9] proposed an LML system based on the multi-task learning developed by Kumar et al. [10]. Here, the learning tasks are autonomous and distributed. In the area of lifelong Unsupervised Learning, Zhiyuan et al. [11] and Wang et al. [12] proposed various lifelong modeling techniques to generate topics from a set of historical tasks and use the past knowledge to develop better topics. A notable application area like the item recommender system using LML emerged [13]. In the field of lifelong Semi-Supervised Learning, a Never-Ending Language Learner is proposed by Carlson et al. [14] and Mitchell et al. [15]. In this approach, through the continuous web crawling a large volume of information is gathered representing entities and relations. A testing scheme for the LML system is proposed by Lianghao et al. [16], where the incremental learning ability of LML is tested to verify the system becomes gradually more knowledgeable over time through accumulation, transfer of knowledge in each iteration. Leveraging the previous research in the area of LML, our model uses an Incremental Lifelong Learning (ILML) approach through Spark Streaming which initializes itself with batch offset at the starting.
We leverage Spark Streaming APIs in our MALA architecture for lifelong learning architecture. Spark Streaming provides an abstraction on streaming datasets called DStreams, or Discretized Streams. DStream is a sequence of data arriving over time [17, 18]. Internally, each DStream is represented as a sequence of RDDs arriving at each time step (hence the name discretized. DStreams can be created from various input sources, such as Flume, Kafka, or HDFS. Once built, they offer two types of operations: transformations, which yield a new DStream, and output operations, which write data to an external system. DStreams provide many of the same operations available on RDDs and additionally provide new operations related to time, such as sliding windows [19]. Spark MLlib library includes APIs for sliding window-based clustering on streaming data.
Datastream clustering widely researched and improved over the years. Among the early works in this area, Guha et al. [20] proposed the STREAM algorithm which produces a constant factor approximation for the k-Median problem in a single pass and using small space. Gupta et al. [21] present a study for outlier detection approaches and case studies for different forms of temporal data. A time decay function that puts variable weights decreasing over time while updating the model is suggested. Our work extends the STREAM algorithm with an enhancement that allows further merging of a large static historical data pool with the latest and most updated streaming model.
Various improvement techniques were proposed for dimension reduction in large scale high-dimensional data. Agarwal et al. [22] present a dimension reduction process for the online learning component where only the limited parameters are learned online and remaining item features are learned through an offline batch process. The model is proved through a recommender system implementation for Yahoo! front page and My Yahoo!. We leverage this approach for the LML system. Apache Spark MLlib implements Random Decision Forest (RDF) for APIs supporting binary and multiclass classification and also for regression models. APIs can handle both numerical and categorical features extending existing MLlib Decision Tree APIs [23]. However, the APIs are computationally infeasible for repeated model re-training with high dimensional data in a streaming setting. To improve the efficiency of the Random Decision Forest algorithm and speed up model updation, we propose online dimension reduction and data sampling technique on Spark DStreams which improves each re-training time by reducing the volume of the training dataset.
This paper is the continuation of our previous work on hybrid incremental learning. As a precursor to the current work [24], first introduces the concept of MALA in the context of Lifelong Learning architecture. Pal et al. [25] further improves the MALA architecture through a novel scheme for reasoning and root cause analysis, streaming hybrid clustering, dimension reduction techniques and in-memory processing capabilities.
Case description
Application scenario
The National Cancer Institute’s Genomic Data Commons (GDC) releases the unified data repository for the cancer research community in support of personalized medicine. The repository contains over 32,000 patient cases including clinical data, treatment data, biopsy results, gene expression data, as well as a whole host of other information [26]. This allows accessibility to researchers who want to uncover novel biomarkers and find a correlation between genes and survival.
Personalized cancer medicine is customized to each individual patient’s need for chemotherapy or drugs based on patients’ specific set of DNA or genes. A cancer patient’s pathological tests like blood, DNA, urine or tissue analysis provide a unique signature based on the DNA combinations. Our analysis enables personalized and targeted medications and achieves a therapeutic response based on the thousands of genotype and phenotype parameters.
Data metadata and model training
The patient repository contains over 32,000 patient cases including clinical data, treatment data, biopsy results, gene expression data, as well as a whole host of other information. This allows us to uncover novel biomarkers, and find a correlation between genes and survival. Each patient data comprises thousands of biometric data for each of the patient’s DNA/RNA sequencing, gene expression, molecular profile, mutational status, drug therapies, survival length, physical characteristics, etc. There are about 21,000 parameters associated with each of the patients, the majority of which is sourced from gene expression data (> 20,500) and the remaining are other pathological identifiers (\(>100\)). For example, primary human genes influencing prostate cancer patient’s overall survival rates are gene HOXB13, MSMB, and CDH1 and the gene expression level in each patient varies with MSMB gene expression level differs between 6 and 15 among all patients in the database. Refer Fig. 3.
The repository contains over 32,000 patient cases including clinical data, treatment data, biopsy results, gene expression data, as well as a whole host of other information. There are about 21,000 parameters associated with each of the patients, majority of which is coming from gene expression data (>20,500) and remaining are other pathological identifiers (\({>}100\)). The objective is to improve overall survival days through root cause analysis and personalized medicine
Our model selects the priority of each feature from about 20,000 genotype and phenotype parameters linked with each record in our training dataset and eliminates a significant number of unrelated parameters. Important features as extracted by the model are listed in below Tables 1 and 2 .
In our model, we create a truly lifelong learning mechanism by dividing the dataset as 50% for batch data at the initialization and remaining data is ingested at a never-ending loop of 30 s sliding window interval. Training and test data volume ratio is kept 80% to 20% with one row relates to one patient records and columns areas mentioned in Tables 1 and 2. The proposed system aims to improve overall survival days by correlating the factors that influence the survivability most. Data is first clustered by a hybrid model of batch and stream. The clustering step isolates the target data points which then proceeds through a three steps process of sampling, feature selection and dimension reduction to built a Decision Tree for reasoning and root cause analysis. Future elements are predicted through Decision Tree Classifier and Regressor for categorical and numerical fields.
Proposed ensemble framework: MALA
The proposed system is based on successive streaming clustering and Random Decision Forest Regressor and Classifier process which uses MALA, a Multi-agent ensemble framework. MALA is described in this section, in the following sections we discuss clustering and Random Decision Forest algorithms.
MALA is a consolidation framework for stream and batch modules based on the fundamentals of LA. MALA is developed as an extension to the standard LA framework and the MALA framework is our main contribution that extends this line of work. The framework itself with batch processing results for streaming engine to incrementally build on top of batch offset. The framework successively applies a streaming clustering procedure followed by a Random Decision Tree algorithm to provide dynamic root cause analysis and forecasting. The framework enables collaborative, accumulative learning through big data tools and APIs. Streaming and batch components act as a cooperative autonomous Multi-agent system. See Fig. 4.
Three layers of MALA consists of the Historical Data Store, stream processor, Knowledge Miner. Real-time and batch layers act as autonomous Multi-agent systems in collaboration. A Multi-agent decision maker component is placed in the MALA stack at the gateway of the data pipeline for further processing
Historical Data Store
A large volume static data pool gets ingested, all at once and at a periodic interval, then is processed and written back, using frameworks like Apache Hadoop and Spark. Input data is stored over time in a distributed file system like HDFS, NoSQL databases or Amazon Simple Storage Service (S3). The model is trained against the entire data pool. In batch mode, the response time is not a big constraint but rather a design is inclined towards a comprehensive coverage of the data. Historical Data Store (HDS) persists the trained models.
Stream processor
The MALA updates its model on each new wave of incoming data. The streaming model initializes itself with saved learning from the batch by loading the trained model from persistent distributed storage into distributed memory. The continuous data stream updates its model incrementally. The training time largely depends on the mini-batch data size and window length. The duration can vary from a few milliseconds to hours. We use Apache Spark Streaming to create in-memory DStreams from the stored model in HDFS produced by the batch jobs. After each iteration of re-training, the updated model gets persisted into memory and disk. Since the model keeps getting larger, the most recent data is cached into distributed memory and the remaining goes to disk. The amount of distributed memory size is configurable through the Spark configuration file. Stream processing includes filtration of data rows and converting, transforming the ingested flow into structured data.
Knowledge Miner and Knowledge Base
Knowledge Miner (KM) consolidates the past learning with recent stream updates into Knowledge Base (KB). KM is responsible for filtration (eliminating unwanted and faulty records), knowledge aggregation and data governance for monitoring and reporting purposes. Filtration and aggregation logic is ad-hoc developed for a specific case study. In our implementation, KM schedules the Spark batch jobs which create the initial training model spawns the Spark streaming clustering jobs to iteratively re-train the model. KM builds the Decision Tree from JSON formatted data provides visualizations and predictive analytics results by ad-hoc query through a web-based search interface. HDFS is used as the persistent storage option for Knowledge Base.
Streaming incremental learning framework
Successive streaming clustering and a Random Decision Forest Regressor and Classifier use a MALA ensemble framework (discussed in preceding section). The system gets its initial offset (or the starting point) through the batch operation. Thereafter, each mini-batch window of stream data needs to re-train the model iteratively. As the data volume grows over time, re-training the entire system on each streaming window becomes computationally infeasible. The proposed framework therefore selectively updates the training model through a dimension reduction method avoiding re-training the entire system.
Problem statement (Cumulative Hybrid Learning): At time t, the system maintains a set of cluster \(C^t = {C_1}, {C_2},..,{C_n}\) with training model \(M^t\) from a past dataset. Once enough training data is collected for a mini-batch window interval \(\delta _t\), \(M^t\) is updated to cover new training model \(M^{t+1}\). Our objective is to build \(M^{t+1}\) with minimum latency by selectively re-training a specific cluster.
(Lifelong Machine Learning (LML) through MALA Architecture: At any point of time t, the learning system has performed n training tasks \(\tau _1, \tau _2,...,\tau _n\) on past datasets \({\mathfrak {D}}_1,{{\mathfrak {D}}_2},...,{{\mathfrak {D}}_n}\). On arrival of the new streaming set \({\mathfrak {D}}_{n+1}\) the training system uses the past acquired knowledge stored in the knowledge base to help learn the current task \(\tau _{n+1}.\)
After accumulation of knowledge \(\tau _{n+1}\), the Knowledge Base is iteratively updated (Fig. 4). The updating involves selective re-training, remove inconsistency, and updating metadata. The learning system updates the current model \(M^t\) to \(M^{t+1}\), that can cluster new data \({\mathfrak {D}}_{n+1}\) into an existing old cluster or form an unseen new cluster. The steps involved are summarized as below:
- 1.
Searching for the similar cluster in \(C^{t}\) that is similar to new dataset \({\mathfrak {D}}_{n+1}\). Create a separate cluster from the clusters in \(C^{t}\).
- 2.
An incremental clustering procedure groups the dataset through the dimension reduction method (“Streaming clustering step” section).
- 3.
The Random Decision Forest algorithm further drills down into each group for comparison between groups and provides reasoning through a Decision Tree graph and predicts each future elements (“Decision tree and Random Decision Forest step” section).
Streaming clustering step
Under the streaming setting, in the bounded small space S, data is accessed through a linear scan (i.e. only once and viewed in order). Established methods for producing a reasonable streaming learning algorithm can be applied here including [27, 28]. The batch version of the k-means algorithm provides an offset (or initial point) for the streaming learning to update the model iteratively.
Algorithm:
- 1.
Divide S into n disparate pieces \(a_1, \ldots,\)\(a_n\).
- 2.
For each i, find \({\mathscr{O}}(k)\) centers in \(a_i\), allocate each element in \(a_i\) to their closest cluster center according to the Euclidean distance function.
- 3.
Let C is the \({\mathscr{O}}(nk)\) center computed in (2), while each center c \(\epsilon \) C is weighted by a number of points allocated to it.
- 4.
Cluster C to get k number of cluster
- 5.
For every streaming window, re-compute new cluster centers using:
$$\begin{aligned} c_{t+1}&= \frac{c_{t~ }n_{t } \alpha +x_{t}m_{t}}{n_{t} \alpha +m_{t}} \end{aligned}$$(1)$$\begin{aligned} n_{t+1}&= n_{t} + m_{t} \end{aligned}$$(2)where \(n_{t }\) is the historical data set and \(c_{t}\) is the old cluster centroid. \(m_{t}\) is the newest data points and \(x_{t}\) is the cluster center computed with new data points. \(\alpha \) is the decay factor. With \(\alpha=0 \), only the latest window dataset is used; for \(\alpha=1 \), an entire historical dataset is used.
Decision tree and Random Decision Forest step
A collection of Decision Trees is generalized to form a more powerful algorithm as Random Decision Forests. The flexibility of Random Decision Forests makes these worthwhile to examine in our dataset. For the number of n features in the dataset, there is \(2^n-2\) possible decision rule (all subset except the empty set and entire set). This leads to the possibility of creating a few billion decision rules for even moderate size data. These decision rules use several heuristics to select the top n decision rule as described below in the steps for constructing a Random Decision Forest:
Step 1. Sampling training subsets
n subsets are sampled from the raw training dataset S using Spark MLLib stratified sampling methods [29]. providing a fraction of sample size and a random seed. For instance, from the patient dataset, men and women patients can be sampled as gender being the key in sampleByKey() method.
Step 2. Important feature selection
Decide how much each feature contributes to the final prediction through Spark featureImportances API [29]. Find the list of the important features over the sampled dataset in the previous step. For a tree ensemble model, Spark RandomForest.featureImportances [30] finds the importance of each feature from about 20,000 genotype and phenotype features associated with each record in our training dataset and eliminate a significant number of unrelated parameters. The method generalizes the concept of Gini Importance for approximating the independent target variable.
Step 3. Dimension reduction of feature vector
We present a unique dimension reduction method of feature vector enabling quick re-training through MALA architecture discussed in the previous section. With this approach, the batch model creates the offset (or initial point). On each streaming window of the dataset, the streaming model just needs to learn the updated features or corrections during the re-training process. Let \(p_{it}\) be the feature vector of item i at time t and let \(q_{it}\) latent factor vector of item i at time t. Then expected training model M at time t is:
where b and A are the regression weight matrix learned through the offline training process. \(p'_{it}b + p'_{it} A \) provides an initial offset for the online training process. Suppose \(q_{it}\) consists of a \( k \)-dimensional linear subspace extended by the columns of a random projection matrix \( B _{ r \times k }\) \(( r \gg k )\) that is,
The online component only learns a k-dimensional vector \(\delta _i\) To eliminate numerical ill-fitting during computations let us assume, \(\delta _i \sim MVN( O ,\sigma _{\delta }^{2} I )\)
where MVN denotes the multivariate normal distribution [22] and \(\sigma ^{2}\) is the variance of noise in actual data. Marginalizing over \(\delta _i\), it is evident that \(q_{it} \sim MVN( O ,\sigma ^{2} B mathnormal{B}')\). Because \(rank( B ) = k < r \), then the probability distribution in a lower-dimensional subspace extended by the columns of \( B \). It is usually better to go with a model which assumes \(q_{it}=B\delta _i + \epsilon _{jt}\), where \(\epsilon _{jt} \sim MVN( O ,\tau ^{2} I )\) is an error of the model after the \( k \)-dimensional linear projection. The process eliminates the rank inadequacy and the distribution of \(q_{it}\sim MVN( O ,\sigma ^{2} B mathnormal{B}') + \tau ^{2} I )\). In spite of the model’s easy compatibility with Gaussian response, due to the model’s computational overhead to the our current model, we assume \(\tau ^{2}=0\). So, \(q_{it}\sim MVN( O ,\sigma ^{2} B B ')\).
Step 4. Constructing the Decision Tree
The Decision Tree is built using the C4.5 algorithm. Primary improvements with C4.5 over the ID3 algorithm are the pruning technique and the capability of handling continuous attributes, null values, and noisy data [31, 32]. In the building process of the tree, for each set of n important features decided in the previous step, select the feature \(f_i\) with the highest normalized information gain on splitting on the feature as the root. Once a feature \(f_i\) is selected as the root of a node, the remaining nodes create the children by recursively calling C4.5.

Sampling n training subset (line 6): n subsets are sampled from the raw training dataset S using the Spark RDD sampling method. For instance, the dataset can be sampled based on gender, age group or patient genetic type.
Extracting important features (lines 7–9): Find a list of the important features over the sampled dataset. The current training dataset contains around 20,000 genotype and phenotype features associated with each patient. This step reduces the number of features to a few hundred based upon the contribution of features to the interdependent variable and removing insignificant features.
Apply batch computation rule (line 10): The batch computation rule process is applied to the historical dataset to update the Knowledge Base and initializes the stream processing engine.
Kafka message queue, (line 11): Kafka acts as a message queue as the subscriber to the stream processing engine.
Re-train the model (lines 13–15): Spark consumes the Kafka queue as a series of DStreams and re-trains the model with the dimension-reduced feature vector. The Knowledge Base is updated subsequently.
Persist the model (line 17): Updated model and Knowledge Base is persisted into HDFS.
Implementing Random Decision Forest through Spark APIs
The Random Decision Forest prediction is based on the weighted average of each tree’s predicted values. For our current dataset, the Random Decision Forest prediction is based on the weighted average on most probable values from each tree and computing the most likely value out of it. On the implementation level, the Spark RandomForestClassifier API is used to build the Random Decision Forest model for categorical features. We have used Spark RandomForestRegressor APIs for numerical features. See “Discussion and evaluation” section for detailed results for numerical and categorical features. The algorithm requires all feature vector to be collected into one column. We use VentorAssembler as a DataFrame transformer within the Spark MlLib pipelines API to build the initial Decision Tree feature vector as the training dataset. The classifier model’s return type is DecisionTreeClassificationModel as MLLib API transforms the data frame and the return value contains predictions as objects. A MulticlassClassificationEvaluator computes the accuracy of classifiers with the F1 score representing a weighted average of precision and recall. We also produce a confusion matrix for the quality of the classifier’s output. A confusion matrix is \(n \times n\) where n is the number of possible outcomes. Values across the rows representing real values while predicted values are placed along columns. The entry at row i and column j represent a count the number of times an instance with true category i was predicted as category j. So, the counts for the correct predictions placed along with the diagonal cells, counts for incorrect predictions are placed in the rest of the cells. See Fig. 5.
Tuning Decision Tree hyperparameters
The following model hyperparameters are tuned to fetch the optimal result: maximum depth, maximum bins, impurity measure, and minimum information gain. Maximum depth is the most number of connected decisions the classifier takes. The value for maximum depth is selected after several trials run on the training set to prevent model overfitting. Bin size is proportional to cluster size with a large bin size leading to an optimal decision rule. The model uses Gini impurity as impurity measure which is directly related to the accuracy of the random-guess classifier measured through the following equation:
where \(p_i\) is the proportion of elements of class i out of N available classes. Minimum Information Gain helps to reject candidate decision rules that do not improve the subsets’ impurity thus resisting model overfitting. We try a number of combinations of the hyperparameters and report the results. The model is trained and evaluated with two values of each hyperparameters producing 16 models in combination. Each model is evaluated by MulticlassClassificationEvaluator in Spark MlLib API [33] to compute the evaluation results for each of the combination of model hyperparameters in terms of accuracy and F1 Score. The objective is o find the optimal values for maximum depth and maximum bins which are just sufficient but not excessive to improve the training time and could resist overfitting. getEstimatorParamMaps [6] API provides accuracy of each combinations of hyperparameters through a single run. This lets us avoid potential thousands of independent test runs for each combination. Note, CrossValidator APIs can also be used for n-fold cross-validations. But it also induces n times more computation complexity and unsuitable for big data applications. The results for the tests select the best combinations for model hyperparameters. The results reveal the Gini impurity works best at a max depth of 30, along with 55 bins. See Fig. 6 showing the sample screenshot of the tests.
Discussion and evaluation
In this section, we present results for experiments with GDC cancer patient data. We start with clustering results that pivot the focus area through outliers. Random Forest results then drills down further to trace the possible consequences leading to a specific outcome. we provide a comparative study about the model performance against the standard machine learning algorithms predicting categorical and numerical features. We also mention probable limitations of the model and alternative approaches.
Experiment setup
The server setup is shown in Table 3. The database for the test was Cassandra Datastax Enterprise Edition (DSE) with a replication factor of 3. All nodes are installed with Cloudera Distribution of Hadoop (CDH) 6.1 and Apache Spark 2.3.1. Algorithms are developed with Scala 2.12.6. Spark adds up the computational power of each individual resource. The total cluster resource is 100 cores, 375 GB memory, 25 TB of HDD. Each node is running 2 Spark executors in the Hadoop YARN cluster mode. Setting the number of executors per node is crucial to achieving optimal parallelism. Small executors will decrease CPU and memory efficiency and big executors will introduce a heavy GC overhead. Spark is run on the YARN cluster mode where the Hadoop Resource Manager performs the role of Spark master and Node Manager work as executor nodes. A Spark Driver running in YARN mode submits an application to the Resource Manager and the Resource Manager designates an Application Master for the Spark application.
Results with GDC cancer patient data
Clustering results
Our database is composed of real-world patient data diagnosed with prostate cancer. 50% of the data is used as historical items and remaining data is streamed at a rate of 10 records per 30 s sliding window interval to re-train the model iteratively. Visualizations are rendered through Splunk Machine Learning Toolkit [34]. We base our Spark API based algorithms through the Python for Scientific Computing add-on in Splunkbase [35].
First, the data is clustered for patient profiling and detecting the outliers. Outliers are further analyzed with a Decision Tree for root cause analysis with the ultimate objective to improve the Overall Surviving (XOS) rate. Cluster charts for first biochemical recurrence and Gleason Score with XOS are shown in Figs. 7 and 8.
Cluster chart for first biochemical recurrence (daysXtoXfirstXbiochemicalXrecurrence) with overall survival (XOS) in days in 3 clusters. In this matrix representation of 4 charts, daysXtoXfirstXbiochemicalXrecurrence and XOS (along x-axis) are plotted with each other and itself along the y-axis. Outliers are further analyzed with a Decision Tree for root cause analysis
Cluster chart for Gleason Score (gleasonXscore) with overall survival (XOS) in days in 3 clusters. In this matrix representation of 4 charts, gleasonXscore and XOS (along x-axis) are plotted with each other and itself along the y-axis. Outliers are further analyzed with a Decision Tree for root cause analysis
The clustering step also detects outliers through distance from the mean computed by the standard deviation (Figs. 9 and 10).
Outliers chart for anomalous Overall Survival (XOS) data. Total 23 outliers are detected (yellow dots) which are further analyzed with Decision Tree for Root Cause Analysis(RCA). Anomalous values are detected through distance from the mean as shown in Fig. 10
The cluster plot reveals that gene expression level along with the Gleason score has a strong correlation to be diagnosed with prostate cancer but no correlation to overall survival days. Figs. 11 and 12 shows the overall survival by target gene TP53 when the Gleason score is categorized into 3 groups (6–7, 8, and 9–10).
Overall survival by cancer causing gene TP53TG5 along with Gleason score clustered into 3 groups (6–7, 8, and 9–10). The data is clustered by proposed hybrid streaming clustering step discussed in “Streaming clustering step” section. The plot shows strong possibility of being diagnosed with prostate cancer with a gene TP53TG5 expression level of 0.5 to 1.5 and Gleason score between 6 to 7
Overall survival by cancer-causing gene TP53TG35 along with Gleason scores clustered into 3 groups (6–7, 8, and 9–10). The data is clustered by the proposed hybrid streaming clustering step discussed in “Streaming clustering step” section. Prostate cancer patients have a much higher probability of TP53TG35 gene expression level between 0.5 to 3 with a Gleason score between 6 to 7
We remark that the clustering methods provide an efficient and actionable grouping and detect the outlier for the patient profiles. Subsequently, the anomalous behavior is correctly summarized in the outlier chart. The efficiency of the process gets improved over time with the incremental lifelong learning approach. Figure 10 shows an explanation for outlier detection. Therefore, in summary, the prototype is said to provide a robust estimation.
Random Decision Forest results
The Random Decision Forest (RDF) Regressor graph further illustrates possible consequences leading to a specific Overall Survival value. Figure 13 shows the Decision Tree splitting process for the portion of the tree. From the tree, it appears that there are significantly contributing genotypes, phenotypes, and other clinical parameters. For instance, days to first biochemical recurrence is an influencing attribute clearly dividing the data pool. A similar conclusion is derived for genes like KFF6 or LRP2.
The Decision Tree split process. Part of the tree is displayed here. The information gain for each feature variable is calculated. The feature with the highest information gain is chosen as the root and splitting node. In each node, information gain is shown as a value along with impurity level which is measured after each split. The process is iterated until the leaf nodes are generated
Figure 14 provides a comparison between influent human genes contributing towards lower Overall Survival (XOS) in days. A Random Decision Forest classifier detects the commonly present genes among prostate cancer patients, this chart facilitates further breakdown by ranking among the available gene pool.
Random Decision Forest provides reasoning through the Decision Tree graph to do the root cause analysis for every anomalous cluster detected by the previous clustering step. We observe the overall survival rate positively influenced if Gleason score \(<7\), if the sample type is not the primary tumor, the patient did not receive radiation therapy and days to biochemical recurrence \(>200\) as identified by the red lines in Fig. 15.
Random Decision Forest drill down for the anomalous cluster. The Decision Tree provides reasoning for root cause analysis. The red lines in the Decision Tree show the positive influence of the Gleason score (\(<7\)), sample type (not a primary tumor), no radiation therapy and days to biochemical recurrence (\(>200\)) on overall survival rate (XOS)
The Decision Tree is shown in Fig. 15 and it accurately indicates the Gleason score as a contributing factor for the overall survival days (see red lines). Figure 16 by the Random Decision Forest algorithm further proves a Gleason score between 6 and 9 has a much lower survival rate of fewer than 1000 days. Finally, a Kaplan–Meier estimator plot [36, 37] is presented based on the Random Decision Forest algorithm. The estimator of survival is given by Eq. 6.
where \(d_i\) and \(n_i\) are the number of deaths and survival at time \(t_1\). See Fig. 17.
Model comparisons
Several experiments are conducted comparing the proposed MALA model with a Decision Tree Regressor, Kernel Ridge, Elastic Net, Ridge, Linear Regression and Lasso models for numeric field predictions. Models are compared in terms of \(R^2\) statistics and RMSE. In general, higher \(R^2\) and lower RMSE signify the model fitting the data better. Results are presented in Table 4. Model prediction accuracy for categorical fields is presented in Table 5. In categorical forecasting, the proposed MALA model is compared with GaussianNB, Bernoulie NB, Decision Tree Classifier, SVM and Logistic Regression in terms of Precision, Recall, Accuracy, and F1 Score. A good model is supposed to exhibit higher values for all comparison parameters. The split for training and test dataset was 50% for both the experiments. Since MALA uses both batch and stream data to fit its model, 50% of the training dataset is used to train at batch setting and the remaining used at streaming setting.
We report actual vs. predicted Line Chart and Scatter Chart (Fig. 18), Residuals Line Chart and Residuals Error Histogram (Fig. 19) for comparative models. Lower values in the Residuals Line Chart and Residuals Error Histogram should prove good model predictions.
The classification accuracy for MALA is 4% greater than SVM, 29% greater that Bernoulie NB and more than 30% greater than all other algorithms. In terms of regression RMSE, the proposed model is 4% better than the Elastic Net, 37% better than the Decision Tree Regressor and more than 40% better than the rest of the algorithms. Furthermore, due to the lifelong incremental learning capabilities, the proposed model significantly improves over time without adding latency in training time. Therefore, compared with comparative models, MALA improves classification and regression accuracy significantly for multiple scales and parameters.
Performance comparison of different machine learning models. Actual vs predicted Line Chart and Scatter Chart. Blue and yellow lines or dots show the actual and predicted values. Proposed MALA shows the higher accuracy than remaining models. a Proposed MALA, b decision tree regressor, c lasso, d linear regression, e kernel ridge, f ridge, g elastic net
Results analysis
The proposed framework provides reasoning through dynamic root cause analysis. Thus, the model’s overall success largely depends on the accuracy of reasoning illustrated through the decision graph. For instance, the Gleason score categorized into 3 groups (6–7, 8, 9–10) has a major influence on the overall Survivability (XOS). To verify the claim, we first plot the probability distribution of XOS to identify if XOS follows a normal distribution through the Lilliefors test [38, 39]. See Fig. 20.
The result h is 1 if the test rejects the null hypothesis at the 5% significance level, and 0 otherwise.
from the Lilliefors test, we reject the null hypothesis at the 5% significance level. Therefore, XOS does not satisfy normal distribution.
Given that XOS does not satisfy normal distribution, we should use other methods to find if the Gleason score has an influence on the overall survival. Hence, we choose a Two-sample Kolmogorov–Smirnov test [40, 41] to validate if the Gleason score has an influence on the overall survival rate. Kolomogorov-Smirnov test determines if two one dimensional probability distribution curve differs. The null hypothesis is: Gleason score groups (6–7, 8, 9–10) does not have any influence on the overall survivability. The objective here is to deny the null hypothesis and prove the model’s reasoning favorably. We call:
The Test returns an h and a p-value. If h=1 we reject the hypothesis (Eq. 7), which means the two vectors XOS (10 < Gleason Score < 6) and XOS(group[1/2/3]) have the different distributions as the test is sensitive to location and shape of the cumulative empirical distribution functions of the two samples. A small p-value (typically \(\le \) 0.1) disproves the null hypothesis . The result is shown Table 6 at 10% significance level.
The value of h is a test decision for the null hypothesis if the two groups of data are from the same continuous distribution (no significant difference). The result h is 1 if the test rejects the null hypothesis at the 10% significance level, and 0 otherwise. We report a positive correlation between Gleason score and overall survivability from Table 6 outcome with h = 1 and \(p\le \alpha \) for all 3 groups. A small p-value provides evidence against the null hypothesis with a significant distance between cumulative empirical distribution functions for two samples. The populations may differ in the shape of the distribution, variability or median. However, in our test, the correlation seems to be tenuous with relatively bigger p values.
Limitations of the proposed model
The framework has the limitation of managing the identical code base for batch and stream layer which yields the same results. This code redundancy may lead to a typical code sync problem since changes in one layer (batch or stream) requires to reflect back the changes to another layer. Also, comparing accuracy, our model would be no superior to the batch only model when trained on the same data volume. But Incremental Lifelong Learning framework gradually improves the prediction accuracy over time and with additional data. One of the shortcomings of Kolmogorov–Smirnov test is that it may not be very powerful since it is modeled to be responsive against every possible kind of deviations between two sample distribution functions. As an alternative to Kolmogorov–Smirnov tests, Area Under The Curve (AUC) measure [42, 43] can also adopted to quantify the efficiency of the proposed model in distinguishing different Gleason Score groups.
Conclusion
In this paper, we proposed an ensemble framework of stream and batch data for incremental lifelong learning. The proposed framework successively applies the first streaming clustering technique and then Random Decision Forest Regressor and Classifier to isolate anomalous patient data and provides reasoning through root cause analysis by feature correlations with an aim to improve the overall survival rate for patients with prostate cancer. While the stream clustering technique creates groups of patient profiles, RDF further drills down into each group for comparison and reasoning. The proposed MALA architecture retains the past learned knowledge and transfers to future learning. The model is able to update the dimension-reduced feature set in every streaming sliding-window without requiring to rebuild the entire training set from scratch. With the arrival of new datasets over time, the model quickly re-trains itself and iteratively becomes more knowledgeable. The Random Decision Forest rule provides the root cause analysis through Decision Tree graphs for only on the anomalous cluster extracted by the clustering rule, removing unnecessary data clutter and pivoting the exact problem domain faster. The proposed Decision Tree contains a relatively complex optimization structure, and capable of predicting the profile of future patients. The Decision Tree, however, is erroneous in a few instances for root cause analysis and forecasting. The classification and regression error in the Decision Tree often arises from null entries in patient genotype and phenotype parameters. In the future, we plan to engage oncologists and clinical pathologists along withC work in practice.
Availability of data and materials
The National Cancer Institute’s Genomic Data Commons (GDC) unified data repository for the cancer patient is available online at Government website: https://gdc.cancer.gov/.
Abbreviations
- GDC:
-
Genomic Data Commons
- HDFS:
-
Hadoop Distributed File System
- DStream:
-
Discretized Stream
- LA:
-
Lambda Architecture
- MALA:
-
multi-agent Lambda Architecture
- KM:
-
Knowledge Miner
- KB:
-
Knowledge Base
- RDF:
-
Random Decision Forest
- S:
-
Simple Storage Service
- HDS:
-
Historical Data Store
- LML:
-
Lifelong Machine Learning
- DSE:
-
Datastax Enterprise Edition
- CDH:
-
Cloudera Distribution of Hadoop
- XOS:
-
overall survival
- RCA:
-
Root Cause Analysis
References
Thrun S. Explanation-based neural network learning: a lifelong learning approach. Boston: Kluwer Academic Publishers; 1996.
Silver DL. The parallel transfer of task knowledge using dynamic learning rates based on a measure of relatedness. Connect Sci. 1996;8(2):277–94. https://doi.org/10.1080/095400996116929.
Silver DL, Mercer RE. The task rehearsal method of life-long learning: overcoming impoverished data. In: Cohen R, Spencer B, editors. Advances in artificial intelligence. Berlin: Springer; 2002. p. 90–101.
Silver DL, Poirier R. Sequential consolidation of learned task knowledge. In: Tawfik AY, Goodwin SD, editors. Advances in artificial intelligence. Berlin: Springer; 2004. p. 217–32.
Silver DL, Mason G, Eljabu L. Consolidation using sweep task rehearsal: overcoming the stability-plasticity problem. In: Barbosa D, Milios E, editors. Advances in artificial intelligence. Cham: Springer; 2015. p. 307–22.
Hong X, Wong P, Liu D, Guan S-U, Man KL, Huang X. Lifelong machine learning: outlook and direction. In: Proceedings of the 2nd international conference on big data research. New York: ACM; 2018. p. 76–79.
Hong X, Pal G, Guan S-U, Wong P, Liu D, Man KL, Huang X. Semi-unsupervised lifelong learning for sentiment classification: Less manual data annotation and more self-studying. In: Proceedings of the 2019 3rd high performance computing and cluster technologies conference. HPCCT 2019. New York: ACM; 2019. p. 87–92. https://doi.org/10.1145/3341069.3342992.
Fei G, Wang S, Liu B. Learning cumulatively to become more knowledgeable. In: Proceedings of the 22Nd ACM SIGKDD international conference on knowledge discovery and data mining. KDD ’16. New York: ACM; 2016. p. 1565–1574. https://doi.org/10.1145/2939672.2939835.
Ruvolo P, Eaton E. ELLA: an efficient lifelong learning algorithm. In: Dasgupta S, McAllester D, editors. Proceedings of the 30th international conference on machine learning. Proceedings of machine learning research, vol. 28. Atlanta: PMLR; 2013. p. 507–515. http://proceedings.mlr.press/v28/ruvolo13.html. Accessed 4 June 2019.
Kumar A, Daume III, H. Learning task grouping and overlap in multi-task learning. 2012; arXiv preprint arXiv:1206.6417.
Chen Z, Liu B. Topic modeling using topics from many domains, lifelong learning and big data. In: International conference on machine learning; 2014. p. 703–711.
Wang S, Chen Z, Liu B. Mining aspect-specific opinion using a holistic lifelong topic model. In: Proceedings of the 25th international conference on world wide web; 2016; International World Wide Web Conferences Steering Committee. p. 167–176.
Liu Q, Liu B, Zhang Y, Kim DS, Gao Z. Improving opinion aspect extraction using semantic similarity and aspect associations. Menlo Park: AAAI; 2016. p. 2986–92.
Carlson A, Betteridge J, Wang RC, Hruschka Jr ER, Mitchell TM. Coupled semi-supervised learning for information extraction. In: Proceedings of the third ACM international conference on web search and data mining. New York: ACM; 2010. p. 101–110.
Mitchell T, Cohen W, Hruschka E, Talukdar P, Yang B, Betteridge J, Carlson A, Dalvi B, Gardner M, Kisiel B, et al. Never-ending learning. Commun ACM. 2018;61(5):103–15.
Li L, Yang Q. Lifelong machine learning test. In: Proceedings of the workshop on “Beyond the Turing Test” of AAAI conference on artificial intelligence; 2015.
Salloum S, Dautov R, Chen X, Peng PX, Huang JZ. Big data analytics on apache spark. Int J Data Sci Anal. 2016;1(3–4):145–64.
Solaimani M, Iftekhar M, Khan L, Thuraisingham B, Ingram JB. Spark-based anomaly detection over multi-source vmware performance data in real-time. In: 2014 IEEE symposium on computational intelligence in cyber security (CICS). New York: IEEE; p. 1–8 2014.
Rettig L, Khayati M, Cudré-Mauroux P, Piórkowski M. Online anomaly detection over big data streams. In: 2015 IEEE international conference on big data (Big Data). New York: IEEE; 2015. p. 1113–1122.
Guha S, Mishra N, Motwani R, O’Callaghan L. Clustering data streams. In: 41st annual symposium On foundations of computer science, 2000. Proceedings. New York: IEEE; 2000. p. 359–366.
Gupta M, Gao J, Aggarwal CC, Han J. Outlier detection for temporal data: a survey. IEEE Trans Knowl Data Eng. 2014;26(9):2250–67.
Agarwal DK, Chen B-C. Statistical methods for recommender systems, Chap. 7. New York: Cambridge University Press; 2016. p. 120–41.
Chen J, Li K, Tang Z, Bilal K, Yu S, Weng C, Li K. A parallel random forest algorithm for big data in a spark cloud computing environment. IEEE Trans Parallel Distrib Syst. 2017;28:919–33.
Pal G, Li G, Atkinson K. Big data ingestion and lifelong learning architecture. In: 2018 IEEE international conference on Big Data (Big Data). New York: IEEE; 2018. p. 5420–5423.
Pal G, Li G, Atkinson K. Multi-agent big-data lambda architecture model for e-commerce analytics. Data. 2018;3(4):58.
https://gdc.cancer.gov/. Accessed 1 June 2019.
https://spark.apache.org/docs/latest/mllib-clustering.html. Accessed 27 Oct 2018.
Guha S, Meyerson A, Mishra N, Motwani R, O’Callaghan L. Clustering data streams: theory and practice. IEEE Trans Knowl Data Eng. 2003;15(3):515–28.
https://spark.apache.org/docs/2.2.0/mllib-statistics.html#stratified-sampling. Accessed 22 Jan 2019.
https://spark.apache.org/docs/2.2.0/api/java/org/apache/spark/ml/classification/RandomForestClassificationModel.html. Accessed 22 Jan 2019.
Hssina B, Merbouha A, Ezzikouri H, Erritali M. A comparative study of decision tree id3 and c4.5. Int J Adv Comput Sci Appl. 2014;. https://doi.org/10.14569/SpecialIssue.2014.040203.
Ruggieri S. Efficient c4.5 [classification algorithm]. IEEE Trans Knowl Data Eng. 2002;14(2):438–44.
https://spark.apache.org/docs/2.2.0/api/java/org/apache/spark/ml/evaluation/MulticlassClassificationEvaluator.html. Accessed 22 Jan 2019.
https://splunkbase.splunk.com/app/2890/. Accessed 2 Feb 2019.
https://splunkbase.splunk.com/. Accessed 2 Feb 2019.
Bland JM, Altman DG. Survival probabilities (the kaplan-meier method). BMJ. 1998;317(7172):1572–80.
Peterson AV Jr. Expressing the kaplan-meier estimator as a function of empirical subsurvival functions. J Am Stat Assoc. 1977;72(360a):854–8.
Razali NM, Wah YB, et al. Power comparisons of shapiro-wilk, Kolmogorov–Smirnov, lilliefors and anderson-darling tests. J Stat Model Anal. 2011;2(1):21–33.
Abdi H, Molin P. Lilliefors/van soest’s test of normality. In: Salkind NJ, Rasmussen K, editors. Encyclopedia of measurement and statistics. Thousand Oaks: Sage; 2007. p. 540–4.
Lilliefors HW. On the Kolmogorov–Smirnov test for normality with mean and variance unknown. J Am Stat Assoc. 1967;62(318):399–402.
Massey FJ Jr. The Kolmogorov–Smirnov test for goodness of fit. J Am Stat Assoc. 1951;46(253):68–78.
Davis J, Goadrich M. The relationship between precision-recall and roc curves. In: Proceedings of the 23rd international conference on machine learning. New York: ACM; 2006. p. 233–240.
Purves RD. Optimum numerical integration methods for estimation of area-under-the-curve (auc) and area-under-the-moment-curve (aumc). J Pharm Biopharm. 1992;20(3):211–26.
Acknowlegements
The authors thank the anonymous reviewers for their helpful suggestions and comments.
Funding
This research was funded by Accenture Technology Labs, Beijing, China. Grant number RDF 15–02–35, Project Code: RDS10120180003.
Author information
Authors and Affiliations
Contributions
All mentioned authors contribute to the elaboration of the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Pal, G., Hong, X., Wang, Z. et al. Lifelong Machine Learning and root cause analysis for large-scale cancer patient data. J Big Data 6, 108 (2019). https://doi.org/10.1186/s40537-019-0261-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40537-019-0261-9