Abstract
Hepatocellular carcinoma (HCC), the most common form of primary liver cancer, typically develops on the background of chronic liver disease and is an aggressive disease with dismal prognosis. Studies using next-generation sequencing of multiple regions of the same tumour nodule suggest different patterns of HCC evolution and confirm the high molecular heterogeneity in a subset of patients. Different hypotheses have been proposed to explain tumour evolution, including clonal selection or neutral and punctuated acquisition of genetic alterations. In parallel, data indicate a fundamental contribution of nonmalignant cells of the tumour microenvironment to cancer clonal evolution. Delineating these dynamics is crucial to improve the treatment of patients with HCC, and particularly to help understand how HCC evolution drives resistance to systemic therapies. A number of new minimally invasive techniques, such as liquid biopsies, could help track cancer evolution in HCC. These tools might improve our understanding of how systemic therapies affect tumour clonal composition and could facilitate implementation of real-time molecular monitoring of patients with HCC.
Key points
-
Hepatocellular carcinoma (HCC), the most frequent form of liver cancer, is a deadly disease that normally arises on the background of chronic liver disease (for example, cirrhosis).
-
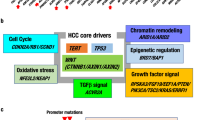
Molecular studies have identified the most frequent alterations of HCC; these include mutations in the TERT promoter, TP53, and CTNNB1, copy number variations, and aberrations in DNA methylation.
-
Large studies on tumour evolution in HCC are lacking, but knowledge can be inferred from small sample studies in HCC utilizing multiregional or longitudinal sampling, including single-cell analysis, and from other tumour entities.
-
Potential drivers of tumour evolution are divided into intrinsic factors (that is, endogenous causes of DNA damage within the evolving tumour cell, such as genomic instability) and extrinsic factors from within the tumour microenvironment, such as immune cells (immunoediting of the tumour) and cancer-associated fibroblasts (remodelling).
-
Clonal tumour composition changes over time might be treatment-induced, and tumour evolution has wide clinical implications. For example, higher intratumoural heterogeneity is correlated with worse clinical outcomes.
-
For the implementation of personalized medicine and to truly understand the clinical effect of tumour evolution in HCC, noninvasive tracing of tumour evolution over time will be of fundamental importance.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Villanueva, A. Hepatocellular carcinoma. N. Engl. J. Med. 380, 1450–1462 (2019).
CDC, National Center for Health Statistics. Trends in liver cancer mortality among adults aged 25 and over in the United States, 2000–2016. CDC, https://www.cdc.gov/nchs/products/databriefs/db314.htm.
Zheng, R. et al. Liver cancer incidence and mortality in China: temporal trends and projections to 2030. Chin. J. Cancer Res. 30, 571–579 (2018).
Nault, J.-C. & Villanueva, A. Intratumor molecular and phenotypic diversity in hepatocellular carcinoma. Clin. Cancer Res. 21, 1786–1788 (2015).
Villanueva, A. et al. Combining clinical, pathology, and gene expression data to predict recurrence of hepatocellular carcinoma. Gastroenterology 140, 1501–1512.e2 (2011).
Hoshida, Y. et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 69, 7385–7392 (2009).
Torrecilla, S. et al. Trunk mutational events present minimal intra- and inter-tumoral heterogeneity in hepatocellular carcinoma. J. Hepatol. 67, 1222–1231 (2017).
Kenmochi, K., Sugihara, S. & Kojiro, M. Relationship of histologic grade of hepatocellular carcinoma (HCC) to tumor size, and demonstration of tumor cells of multiple different grades in single small HCC. Liver 7, 18–26 (2008).
Nowell, P. C. The clonal evolution of tumor cell populations. Science 194, 23–28 (1976).
Xue, R. et al. Variable intra-tumor genomic heterogeneity of multiple lesions in patients with hepatocellular carcinoma. Gastroenterology 150, 998–1008 (2016).
Gerlinger, M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 (2012).
Yachida, S. et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 467, 1114–1117 (2010).
Sottoriva, A. et al. A Big Bang model of human colorectal tumor growth. Nat. Genet. 47, 209–216 (2015).
Ling, S. et al. Extremely high genetic diversity in a single tumor points to prevalence of non-Darwinian cell evolution. Proc. Natl. Acad. Sci. USA. E6496–E6505 (2015).
Williams, M. J., Werner, B., Barnes, C. P., Graham, T. A. & Sottoriva, A. Identification of neutral tumor evolution across cancer types. Nat. Genet. 48, 238–244 (2016).
Zucman-Rossi, J., Villanueva, A., Nault, J.-C. & Llovet, J. M. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology 149, 1226–1239.e4 (2015).
Nault, J. C. et al. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat. Commun. 4, 2218 (2013).
Iavarone, M. et al. Contrast enhanced CT-scan to diagnose intrahepatic cholangiocarcinoma in patients with cirrhosis. J. Hepatol. 58, 1188–1193 (2013).
Di Tommaso, L. et al. Advanced precancerous lesions in the liver. Best Pract. Res. Clin. Gastroenterol. 27, 269–284 (2013).
Nault, J. C. et al. Telomerase reverse transcriptase promoter mutation is an early somatic genetic alteration in the transformation of premalignant nodules in hepatocellular carcinoma on cirrhosis. Hepatology 60, 1983–1992 (2014).
Nault, J.-C., Ningarhari, M., Rebouissou, S. & Zucman-Rossi, J. The role of telomeres and telomerase in cirrhosis and liver cancer. Nat. Rev. Gastroenterol. Hepatol. 16, 544–558 (2019).
Farazi, P. A. et al. Differential impact of telomere dysfunction on initiation and progression of hepatocellular carcinoma. Cancer Res. 63, 5021–5027 (2003).
Günes, C. & Rudolph, K. L. The role of telomeres in stem cells and cancer. Cell 152, 390–393 (2013).
Zhu, M. et al. Somatic mutations increase hepatic clonal fitness and regeneration in chronic liver disease. Cell 177, 608–621.e12 (2019).
Um, T.-H. et al. Aberrant CpG island hypermethylation in dysplastic nodules and early HCC of hepatitis B virus-related human multistep hepatocarcinogenesis. J. Hepatol. 54, 939–947 (2011).
Villanueva, A. et al. DNA methylation-based prognosis and epidrivers in hepatocellular carcinoma. Hepatology 61, 1945–1956 (2015).
Nault, J.-C., Bioulac-Sage, P. & Zucman-Rossi, J. Hepatocellular benign tumors — from molecular classification to personalized clinical care. Gastroenterology 144, 888–902 (2013).
Nault, J.-C., Paradis, V., Cherqui, D., Vilgrain, V. & Zucman-Rossi, J. Molecular classification of hepatocellular adenoma in clinical practice. J. Hepatol. 67, 1074–1083 (2017).
Nault, J.-C. et al. Molecular classification of hepatocellular adenoma associates with risk factors, bleeding, and malignant transformation. Gastroenterology 152, 880–894.e6 (2017).
Pilati, C. et al. Genomic profiling of hepatocellular adenomas reveals recurrent FRK-activating mutations and the mechanisms of malignant transformation. Cancer Cell 25, 428–441 (2014).
Bioulac-Sage, P., Sempoux, C. & Balabaud, C. Hepatocellular adenomas: morphology and genomics. Gastroenterol. Clin. North Am. 46, 253–272 (2017).
Rebouissou, S. et al. Genotype-phenotype correlation of CTNNB1 mutations reveals different β-catenin activity associated with liver tumor progression. Hepatology 64, 2047–2061 (2016).
Chen, Y., Wong, P. P., Sjeklocha, L., Steer, C. J. & Sahin, M. B. Mature hepatocytes exhibit unexpected plasticity by direct dedifferentiation into liver progenitor cells in culture. Hepatology 55, 563–574 (2012).
Tarlow, B. D. et al. Bipotential adult liver progenitors are derived from chronically injured mature hepatocytes. Cell Stem Cell 15, 605–618 (2014).
Seehawer, M. et al. Necroptosis microenvironment directs lineage commitment in liver cancer. Nature 562, 69–75 (2018).
Sia, D., Villanueva, A., Friedman, S. L. & Llovet, J. M. Liver cancer cell of origin, molecular class, and effects on patient prognosis. Gastroenterology 152, 745–761 (2017).
Castelli, G., Pelosi, E. & Testa, U. Liver cancer: molecular characterization, clonal evolution and cancer stem cells. Cancers 9, E127 (2017).
Lee, J.-S. et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat. Med. 12, 410–416 (2006).
Brunt, E. et al. cHCC-CCA: consensus terminology for primary liver carcinomas with both hepatocytic and cholangiocytic differentiation. Hepatology 68, 113–126 (2018).
Moeini, A. et al. Mixed hepatocellular cholangiocarcinoma tumors: cholangiolocellular carcinoma is a distinct molecular entity. J. Hepatol. 66, 952–961 (2017).
Jeon, J. et al. Comparing clonality between components of combined hepatocellular carcinoma and cholangiocarcinoma by targeted sequencing. Cancer Genomics Proteom. 15, 291–298 (2018).
Stavraka, C., Rush, H. & Ross, P. Combined hepatocellular cholangiocarcinoma (cHCC-CC): an update of genetics, molecular biology, and therapeutic interventions. J. Hepatocell. Carcinoma 6, 11–21 (2019).
Xue, R. et al. Genomic and transcriptomic profiling of combined hepatocellular and intrahepatic cholangiocarcinoma reveals distinct molecular subtypes. Cancer Cell 35, 932–947.e8 (2019).
Wang, A. et al. Whole-exome sequencing reveals the origin and evolution of hepato-cholangiocarcinoma. Nat. Commun. 9, 894 (2018).
Fujimoto, A. et al. Whole-genome mutational landscape of liver cancers displaying biliary phenotype reveals hepatitis impact and molecular diversity. Nat. Commun. 6, 6120 (2015).
Cazals-Hatem, D. et al. Clinical and molecular analysis of combined hepatocellular-cholangiocarcinomas. J. Hepatol. 41, 292–298 (2004).
Joseph, N. M. et al. Genomic profiling of combined hepatocellular–cholangiocarcinoma reveals similar genetics to hepatocellular carcinoma. J. Pathol. 248, 164–178 (2019).
Liu, Z.-H. et al. Whole-exome mutational and transcriptional landscapes of combined hepatocellular cholangiocarcinoma and intrahepatic cholangiocarcinoma reveal molecular diversity. Biochim. Biophys. Acta Mol. Basis Dis. 1864, 2360–2368 (2018).
He, J. et al. Block of NF-kB signaling accelerates MYC-driven hepatocellular carcinogenesis and modifies the tumor phenotype towards combined hepatocellular cholangiocarcinoma. Cancer Lett. 458, 113–122 (2019).
Mishra, L. et al. Liver stem cells and hepatocellular carcinoma. Hepatology 49, 318–329 (2009).
Zheng, H. et al. Single-cell analysis reveals cancer stem cell heterogeneity in hepatocellular carcinoma. Hepatology 68, 127–140 (2018).
Hirata, A., Hatano, Y., Niwa, M., Hara, A. & Tomita, H. Heterogeneity in colorectal cancer stem cells. Cancer Prev. Res. 12, 413–420 (2019).
Hatina, J. et al. Ovarian cancer stem cell heterogeneity. Adv. Exp. Med. Biol. 1139, 201–221 (2019).
Friemel, J. et al. Intratumor heterogeneity in hepatocellular carcinoma. Clin. Cancer Res. 21, 1951–1961 (2015).
Zhai, W. et al. The spatial organization of intra-tumour heterogeneity and evolutionary trajectories of metastases in hepatocellular carcinoma. Nat. Commun. 8, 4565 (2017).
Lin, D.-C. et al. Genomic and epigenomic heterogeneity of hepatocellular carcinoma. Cancer Res. 77, 2255–2265 (2017).
Roerink, S. F. et al. Intra-tumour diversification in colorectal cancer at the single-cell level. Nature 556, 457–462 (2018).
Villani, A.-C. et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356, eaah4573 (2017).
Hou, Y. et al. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res. 26, 304–319 (2016).
Marquardt, J. U., Andersen, J. B. & Thorgeirsson, S. S. Functional and genetic deconstruction of the cellular origin in liver cancer. Nat. Rev. Cancer 15, 653–667 (2015).
Duan, M. et al. Diverse modes of clonal evolution in HBV-related hepatocellular carcinoma revealed by single-cell genome sequencing. Cell Res. 28, 359–373 (2018).
Shi, J.-Y. et al. Inferring the progression of multifocal liver cancer from spatial and temporal genomic heterogeneity. Oncotarget 7, 2867–2877 (2016).
Furuta, M. et al. Whole genome sequencing discriminates hepatocellular carcinoma with intrahepatic metastasis from multi-centric tumors. J. Hepatol. 66, 363–373 (2017).
Reiter, J. G. et al. Minimal functional driver gene heterogeneity among untreated metastases. Science 361, 1033–1037 (2018).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Cimini, D. Merotelic kinetochore orientation, aneuploidy, and cancer. Biochim. Biophys. Acta 1786, 32–40 (2008).
Taylor, A. M. et al. Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell 33, 676–689.e3 (2018).
Bakhoum, S. F. & Landau, D. A. Chromosomal instability as a driver of tumor heterogeneity and evolution. Cold Spring Harb. Perspect. Med. 7, a029611 (2017).
Bakhoum, S. F. & Cantley, L. C. The multifaceted role of chromosomal instability in cancer and its microenvironment. Cell 174, 1347–1360 (2018).
Laughney, A. M., Elizalde, S., Genovese, G. & Bakhoum, S. F. Dynamics of tumor heterogeneity derived from clonal karyotypic evolution. Cell Rep. 12, 809–820 (2015).
de Bruin, E. C. et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 346, 251–256 (2014).
Swanton, C., McGranahan, N., Starrett, G. J. & Harris, R. S. APOBEC enzymes: mutagenic fuel for cancer evolution and heterogeneity. Cancer Discov. 5, 704–712 (2015).
Burns, M. B., Temiz, N. A. & Harris, R. S. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat. Genet. 45, 977–983 (2013).
Raynaud, F., Mina, M., Tavernari, D. & Ciriello, G. Pan-cancer inference of intra-tumor heterogeneity reveals associations with different forms of genomic instability. PLoS Genet. 14, e1007669 (2018).
Yang, Z. et al. Overexpression of APOBEC3F in tumor tissues is potentially predictive for poor recurrence-free survival from HBV-related hepatocellular carcinoma. Discov. Med. 20, 349–356 (2015).
Yang, Z. et al. Correlation of APOBEC3 in tumor tissues with clinico-pathological features and survival from hepatocellular carcinoma after curative hepatectomy. Int. J. Clin. Exp. Med. 8, 7762–7769 (2015).
Xu, R. et al. Association of human APOBEC3 cytidine deaminases with the generation of hepatitis virus B x antigen mutants and hepatocellular carcinoma. Hepatology 46, 1810–1820 (2007).
Mittal, D., Gubin, M. M., Schreiber, R. D. & Smyth, M. J. New insights into cancer immunoediting and its three component phases — elimination, equilibrium and escape. Curr. Opin. Immunol. 27, 16–25 (2014).
Liu, X. S. & Mardis, E. R. Applications of immunogenomics to cancer. Cell 168, 600–612 (2017).
Milo, I. et al. The immune system profoundly restricts intratumor genetic heterogeneity. Sci. Immunol. 3, eaat1435 (2018).
Wolf, Y. et al. UVB-induced tumor heterogeneity diminishes immune response in melanoma. Cell 179, 219–235.e21 (2019).
Gil Del Alcazar, C. R. et al. Immune escape in breast cancer during in situ to invasive carcinoma transition. Cancer Discov. 7, 1098–1115 (2017).
Janiszewska, M. et al. Subclonal cooperation drives metastasis by modulating local and systemic immune microenvironments. Nat. Cell Biol. 21, 879–888 (2019).
Zhang, A. W. et al. Interfaces of malignant and immunologic clonal dynamics in ovarian cancer. Cell 173, 1755–1769.e22 (2018).
Angelova, M. et al. Evolution of metastases in space and time under immune selection. Cell 175, 751–765.e16 (2018).
Nishida, N. & Kudo, M. Immunological microenvironment of hepatocellular carcinoma and its clinical implication. Oncology 92, 40–49 (2017).
Zheng, C. et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell 169, 1342–1356.e16 (2017).
Shi, L. et al. Multi-omics study revealing the complexity and spatial heterogeneity of tumor-infiltrating lymphocytes in primary liver carcinoma. Oncotarget 8, 34844–34857 (2017).
Snyder, A. et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 371, 2189–2199 (2014).
Rizvi, N. A. et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128 (2015).
Pinyol, R., Sia, D. & Llovet, J. M. Immune exclusion-Wnt/CTNNB1 class predicts resistance to immunotherapies in HCC. Clin. Cancer Res. 25, 2021–2023 (2019).
Sia, D. et al. Identification of an immune-specific class of hepatocellular carcinoma, based on molecular features. Gastroenterology 153, 812–826 (2017).
Spranger, S., Bao, R. & Gajewski, T. F. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature 523, 231–235 (2015).
Kubo, N., Araki, K., Kuwano, H. & Shirabe, K. Cancer-associated fibroblasts in hepatocellular carcinoma. World J. Gastroenterol. 22, 6841–6850 (2016).
Jia, C.-C. et al. Cancer-associated fibroblasts from hepatocellular carcinoma promote malignant cell proliferation by HGF secretion. PLoS One 8, e63243 (2013).
Song, T., Dou, C., Jia, Y., Tu, K. & Zheng, X. TIMP-1 activated carcinoma-associated fibroblasts inhibit tumor apoptosis by activating SDF1/CXCR4 signaling in hepatocellular carcinoma. Oncotarget 6, 12061–12079 (2015).
Shirabe, K. et al. Role of tumor-associated macrophages in the progression of hepatocellular carcinoma. Surg. Today 42, 1–7 (2012).
Gorelik, E., Wiltrout, R. H., Brunda, M. J., Holden, H. T. & Herberman, R. B. Augmentation of metastasis formation by thioglycollate-elicited macrophages. Int. J. Cancer 29, 575–581 (1982).
Capece, D. et al. The inflammatory microenvironment in hepatocellular carcinoma: a pivotal role for tumor-associated macrophages. Biomed Res. Int. 2013, 187204 (2013).
Pollard, J. W. Tumour-educated macrophages promote tumour progression and metastasis. Nat. Rev. Cancer 4, 71–78 (2004).
Burrell, R. A. & Swanton, C. Tumour heterogeneity and the evolution of polyclonal drug resistance. Mol. Oncol. 8, 1095–1111 (2014).
Morris, L. G. T. et al. Pan-cancer analysis of intratumor heterogeneity as a prognostic determinant of survival. Oncotarget 7, 10051–10063 (2016).
Yang, F. et al. Intratumor heterogeneity predicts metastasis of triple-negative breast cancer. Carcinogenesis 38, 900–909 (2017).
Jamal-Hanjani, M. et al. Tracking the evolution of non-small-cell lung cancer. N. Engl. J. Med. 376, 2109–2121 (2017).
Reuben, A. et al. TCR repertoire intratumor heterogeneity in localized lung adenocarcinomas: an association with predicted neoantigen heterogeneity and postsurgical recurrence. Cancer Discov. 7, 1088–1097 (2017).
Sharma, P. & Allison, J. P. The future of immune checkpoint therapy. Science 348, 56–61 (2015).
McGranahan, N. et al. Allele-specific HLA loss and immune escape in lung cancer evolution. Cell 171, 1259–1271.e11 (2017).
Yarchoan, M., Hopkins, A. & Jaffee, E. M. Tumor mutational burden and response rate to PD-1 inhibition. N. Engl. J. Med. 377, 2500–2501 (2017).
Le, D. T. et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 372, 2509–2520 (2015).
Le, D. T. et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413 (2017).
FDA. FDA approves first cancer treatment for any solid tumor with a specific genetic feature. FDA, https://www.fda.gov/news-events/press-announcements/fda-approves-first-cancer-treatment-any-solid-tumor-specific-genetic-feature
Salem, M. E. et al. Landscape of tumor mutation load, mismatch repair deficiency, and PD-L1 expression in a large patient cohort of gastrointestinal cancers. Mol. Cancer Res. 16, 805–812 (2018).
Sanchez-Vega, F. et al. Oncogenic signaling pathways in the cancer genome atlas. Cell 173, 321–337.e10 (2018).
Harding, J. J. et al. Prospective genotyping of hepatocellular carcinoma: clinical implications of next generation sequencing for matching patients to targeted and immune therapies. Clin. Cancer Res. 25, 2116–2126 (2018).
Davoli, T., Uno, H., Wooten, E. C. & Elledge, S. J. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 355, eaaf8399 (2017).
Bruix, J. et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 389, 56–66 (2017).
Kudo, M. et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet 391, 1163–1173 (2018).
Llovet, J. M. et al. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 359, 378–390 (2008).
Abou-Alfa, G. K. et al. Cabozantinib in patients with advanced and progressing hepatocellular carcinoma. N. Engl. J. Med. 379, 54–63 (2018).
Zhu, A. X. et al. Ramucirumab after sorafenib in patients with advanced hepatocellular carcinoma and increased α-fetoprotein concentrations (REACH-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 20, 282–296 (2019).
Hu, G. et al. In vivo acquired sorafenib-resistant patient-derived tumor model displays alternative angiogenic pathways, multi-drug resistance and chromosome instability. Oncol. Lett. 16, 3439–3446 (2018).
Firtina Karagonlar, Z., Koc, D., Iscan, E., Erdal, E. & Atabey, N. Elevated hepatocyte growth factor expression as an autocrine c-Met activation mechanism in acquired resistance to sorafenib in hepatocellular carcinoma cells. Cancer Sci. 107, 407–416 (2016).
Tovar, V. et al. Tumour initiating cells and IGF/FGF signalling contribute to sorafenib resistance in hepatocellular carcinoma. Gut 66, 530–540 (2015).
Amirouchene-Angelozzi, N., Swanton, C. & Bardelli, A. Tumor evolution as a therapeutic target. Cancer Discov. https://doi.org/10.1158/2159-8290.CD-17-0343 (2017).
Marrero, J. A. et al. Diagnosis, staging, and management of hepatocellular carcinoma: 2018 practice guidance by the American Association for the Study of Liver Diseases. Hepatology 68, 723–750 (2018).
European Association for the Study of the Liver. EASL clinical practice guidelines: management of hepatocellular carcinoma. J. Hepatol. 69, 182–236 (2018).
Dazert, E. et al. Quantitative proteomics and phosphoproteomics on serial tumor biopsies from a sorafenib-treated HCC patient. Proc. Natl. Acad. Sci. USA 113, 1381–1386 (2016).
Labgaa, I. & Villanueva, A. Liquid biopsy in liver cancer. Discov. Med. 19, 263–273 (2015).
Abbosh, C. et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 545, 446–451 (2017).
Murtaza, M. et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 497, 108–112 (2013).
Wang, D.-S. et al. Liquid biopsies to track trastuzumab resistance in metastatic HER2-positive gastric cancer. Gut 68, 1152–1161 (2018).
Bernard, V. et al. Circulating nucleic acids are associated with outcomes of patients with pancreatic cancer. Gastroenterology 156, 108–118.e4 (2019).
Khan, K. H. et al. Longitudinal liquid biopsy and mathematical modeling of clonal evolution forecast time to treatment failure in the PROSPECT-C phase II colorectal cancer clinical trial. Cancer Discov. 8, 1270–1285 (2018).
Merker, J. D. et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists joint review. J. Clin. Oncol. 36, 1631–1641 (2018).
Labgaa, I. et al. A pilot study of ultra-deep targeted sequencing of plasma DNA identifies driver mutations in hepatocellular carcinoma. Oncogene 37, 3740–3752 (2018).
Huang, A. et al. Detecting circulating tumor DNA in hepatocellular carcinoma patients using droplet digital PCR is feasible and reflects intratumoral heterogeneity. J. Cancer 7, 1907–1914 (2016).
Cai, Z.-X. et al. Circulating tumor DNA profiling reveals clonal evolution and real-time disease progression in advanced hepatocellular carcinoma. Int. J. Cancer 141, 977–985 (2017).
Xu, R.-H. et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat. Mater. 16, 1155–1161 (2017).
Kisiel, J. B. et al. Hepatocellular carcinoma detection by plasma methylated DNA: discovery, phase I pilot, and phase II clinical validation. Hepatology 69, 1180–1192 (2019).
Jiang, P. et al. Preferred end coordinates and somatic variants as signatures of circulating tumor DNA associated with hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 115, E10925–E10933 (2018).
Ogle, L. F. et al. Imagestream detection and characterisation of circulating tumour cells — a liquid biopsy for hepatocellular carcinoma? J. Hepatol. 65, 305–313 (2016).
D’Avola, D. et al. High-density single cell mRNA sequencing to characterize circulating tumor cells in hepatocellular carcinoma. Sci. Rep. 8, 11570 (2018).
Kalinich, M. et al. An RNA-based signature enables high specificity detection of circulating tumor cells in hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 114, 1123–1128 (2017).
Felden, J. von et al. Circulating tumor cells as liquid biomarker for high HCC recurrence risk after curative liver resection. Oncotarget 8, 89978–89987 (2017).
Xue, F. et al. Application of a novel liquid biopsy in patients with hepatocellular carcinoma undergoing liver transplantation. Oncol. Lett. 15, 5481–5488 (2018).
Gatenby, R. A., Grove, O. & Gillies, R. J. Quantitative imaging in cancer evolution and ecology. Radiology 269, 8–15 (2013).
Hectors, S. J. et al. Quantification of hepatocellular carcinoma heterogeneity with multiparametric magnetic resonance imaging. Sci. Rep. 7, 2452 (2017).
Segal, E. et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat. Biotechnol. 25, 675–680 (2007).
Xia, W. et al. Radiogenomics of hepatocellular carcinoma: multiregion analysis-based identification of prognostic imaging biomarkers by integrating gene data — a preliminary study. Phys. Med. Biol. 63, 035044 (2018).
Bensch, F. et al. 89Zr-atezolizumab imaging as a non-invasive approach to assess clinical response to PD-L1 blockade in cancer. Nat. Med. 24, 1852–1858 (2018).
Johnson, P. J. et al. Assessment of liver function in patients with hepatocellular carcinoma: a new evidence-based approach — the ALBI grade. J. Clin. Oncol. 33, 550–558 (2015).
Weinstein, I. B. Cancer. Addiction to oncogenes — the Achilles heal of cancer. Science 297, 63–64 (2002).
Hyman, D. M. et al. Vemurafenib in multiple nonmelanoma cancers with BRAFV600 mutations. N. Engl. J. Med. 373, 726–736 (2015).
Shaw, A. T. et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N. Engl. J. Med. 368, 2385–2394 (2013).
Choi, Y. L. et al. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N. Engl. J. Med. 363, 1734–1739 (2010).
Glickman, M. S. & Sawyers, C. L. Converting cancer therapies into cures: lessons from infectious diseases. Cell 148, 1089–1098 (2012).
Vermehren, J., Park, J. S., Jacobson, I. M. & Zeuzem, S. Challenges and perspectives of direct antivirals for the treatment of hepatitis C virus infection. J. Hepatol. 69, 1178–1187 (2018).
Koeberle, D. et al. Sorafenib with or without everolimus in patients with advanced hepatocellular carcinoma (HCC): a randomized multicenter, multinational phase II trial (SAKK 77/08 and SASL 29). Ann. Oncol. 27, 856–861 (2016).
Martins-Filho, S. N. et al. A phenotypical map of disseminated hepatocellular carcinoma suggests clonal constraints in metastatic sites. Histopathology 74, 718–730 (2019).
Buczak, K. et al. Spatial tissue proteomics quantifies inter- and intratumor heterogeneity in hepatocellular carcinoma (HCC). Mol. Cell. Proteom. 17, 810–825 (2018).
Acknowledgements
The work of A.V. is supported by the U.S. Department of Defense (CA150272P3). The work of J.v.F. is supported by Deutsche Forschungsgemeinschaft (FE1746/1-1). The work of T.G.-L is supported by Asociación Española para el Estudio del Hígado (AEEH).
Author information
Authors and Affiliations
Contributions
A.V. made a substantial contribution to discussion of content, and wrote and reviewed/edited the manuscript before submission. A.J.C., J.v.F., S.S. and T.G.-L. researched data for the article, made a substantial contribution to discussion of content, and wrote and reviewed/edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
A.V. has received consulting fees from Guidepoint and Fujifilm, advisory board fees from Exact Sciences, Nucleix and NGM, and lecture fees from Exelixis. The other authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Craig, A.J., von Felden, J., Garcia-Lezana, T. et al. Tumour evolution in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol 17, 139–152 (2020). https://doi.org/10.1038/s41575-019-0229-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41575-019-0229-4
This article is cited by
-
Identification and validation of a novel predictive signature based on hepatocyte-specific genes in hepatocellular carcinoma by integrated analysis of single-cell and bulk RNA sequencing
BMC Medical Genomics (2024)
-
Identification of fatty acids synthesis and metabolism-related gene signature and prediction of prognostic model in hepatocellular carcinoma
Cancer Cell International (2024)
-
UTP11 promotes the growth of hepatocellular carcinoma by enhancing the mRNA stability of Oct4
BMC Cancer (2024)
-
An exosome mRNA-related gene risk model to evaluate the tumor microenvironment and predict prognosis in hepatocellular carcinoma
BMC Medical Genomics (2024)
-
O-GlcNAcylation of E3 ubiquitin ligase SKP2 promotes hepatocellular carcinoma proliferation
Oncogene (2024)