Abstract
Background
The development of prostate cancer can be influenced by genetic and environmental factors. Numerous germline SNPs influence prostate cancer susceptibility. The functional pathways in which these SNPs increase prostate cancer susceptibility are unknown. Finasteride is currently not being used routinely as a chemoprevention agent but the long term outcomes of the PCPT trial are awaited. The outcomes of the SELECT trial have not recommended the use of chemoprevention in preventing prostate cancer. This study investigated whether germline risk SNPs could be used to predict outcomes in the PCPT and SELECT trial.
Methods
Genotyping was performed in European men entered into the PCPT trial (n = 2434) and SELECT (n = 4885). Next generation genotyping was performed using Affymetrix® Eureka™ Genotyping protocols. Logistic regression models were used to test the association of risk scores and the outcomes in the PCPT and SELECT trials.
Results
Of the 100 SNPs, 98 designed successfully and genotyping was validated for samples genotyped on other platforms. A number of SNPs predicted for aggressive disease in both trials. Men with a higher polygenic score are more likely to develop prostate cancer in both trials, but the score did not predict for other outcomes in the trial.
Conclusion
Men with a higher polygenic risk score are more likely to develop prostate cancer. There were no interactions of these germline risk SNPs and the chemoprevention agents in the SELECT and PCPT trials.
Similar content being viewed by others
Introduction
Prostate cancer accounts for a quarter of cancer diagnoses in men in the UK and is the fourth most common cancer worldwide with an estimated 1.1 million men diagnosed in 2012 [1]. Screening strategies have not led to their routine clinical use in daily practice, due to an over-diagnosis of indolent cancers [2].
Due to its biology, prostate cancer is ideally suited target for chemoprevention because of its step-wise biological development. Prostate cancer may have precursor lesions such as atypical small acinar proliferation and high grade prostatic intraepithelial neoplasia which may appear many years before cancer is diagnosed. This high risk population with these precursor lesions could be targeted for chemoprevention [3]. Prostate cancer also has a long latent period from its development to its eventual clinical manifestation. Chemoprevention could also be used to prevent these early malignant lesions developing further and therefore many men could be spared diagnostic procedures and potentially toxic systemic treatments [4].
It is thought that the risk of prostate cancer development could be potentially modifiable by dietary and other lifestyle factors. Large epidemiological studies have shown that migrants who have moved from areas of low prostate cancer incidence such as Korea and Japan to the USA have developed prostate cancer rates similar to their native inhabitants [5, 6]. Environmental factors are attributable to this change of risk and some of these could be potentially modifiable by dietary changes [6]. There are also causal links between androgen exposure and the development of prostate cancer, which is also potentially modifiable [7]. The need to answer the question of the role of chemoprevention in prostate cancer was the aim of two large randomised controlled trials: The Prostate cancer Prevention Trial (PCPT) and The Selenium and Vitamin E Cancer Prevention Trial (SELECT). The biological rationale for the agents used in the trial, trial schema and results can be found in more detail elsewhere [8, 9]. In summary, 18,880 men were eligible and randomised on the PCPT to receive placebo or finasteride. The results of the PCPT trial showed that prostate cancer was detected in 803 of the 4368 men in the finasteride group and 1147 of the 4692 men in the placebo group who had data for analyses. There was a 24.8% reduction in prevalence over the 7-year period. Tumours with a Gleason score 7–10 were more common in the finasteride group (280 of 757 tumours (37.0%)), than in the placebo group (237 of 1068 tumours (22.2%)) (P < 0.001 for the comparison between groups) [8]. The long-term results of the PCPT trial showed that the 10 year survival from prostate cancer was equivalent between the 2 groups [10].
In the SELECT trial, 35,533 men were randomised to receive selenium or vitamin E, or both or neither. The SELECT trial was closed early because the interim analysis showed that neither selenium, vitamin E, nor the combination prevented prostate cancer. The long-term results of the SELECT trial showed that men taking vitamin E had increased incidence of prostate cancer compared with placebo, which was statistically significant [9]. Genetic variations influencing the effects of these chemoprevention agents have been reported from the PCPT and SELECT trials [11,12,13,14]. However these studies have only used a candidate gene approach to look for association in comparison with our study which has followed up validated risk single nucleotide polymorphisms (SNPs).
At the time of undertaking these analyses, 100 germline risk SNPs had been identified in genome wide association studies [15, 16]. The 100 SNPs account for ~33% of familial risk of prostate cancer [16]. If polygenic risk scores are calculated, the top 10% of men in the highest risk stratum have an ~2.9-fold relative risk of prostate cancer compared with the average of the population, whilst the top 1% of men have a 5.7-fold relative risk in comparison with the population average [16]. If these polygenic SNP scores could be incorporated into screening with the addition of known risk factors (age, race, and family history (FHx)), then men with the highest risk of prostate cancer development could be targeted for more intensive screening or intervention.
Traditional genotyping technologies are currently not suited to provide fast accurate outputs that could be used in prospective clinical trials [17]. Genotype calling from next generation sequence techniques may be able to overcome these issues and therefore be more suited for everyday clinical use [18]. We have worked with Eureka Genomics (Hercules, CA; previously affymetrix now ThermoFisher Scientific) to design and validate a next generation genotyping assay in these analyses.
While the functional role of how these SNPs influence cancer predisposition is under investigation, evidence suggests that this may be through various pathways [19]. Therefore, the hypothesis underlying this study was that common genetic variants involved in prostate cancer predisposition may have a role in tumour formation but also influence the effects of the chemoprevention agents. In breast cancer, evidence has shown that chemoprevention with tamoxifen reduces the risk of contralateral breast cancer for those women who have variants in the BRCA2 gene [20]. The aim of this study was to investigate the association of prostate cancer germline risk SNPs and their influence on the chemoprevention agents in the PCPT and SELECT trials.
Materials and methods
Patients
The details of the PCPT study can be found in more detail elsewhere [8]. Informed consent as obtained from all patients. In summary, healthy men aged 55 years or older, with a normal digital rectal examination (DRE), American Association Urology Score of less than 20 and no clinically significant co-morbidity were entered into the trial. The prostate specific antigen (PSA) on entry into the trial was required to be 3.0 ng/ml or lower. Men were randomised to receive Finasteride or placebo. The men underwent annual DRE, and measurements of PSA. If the DRE was suspicious for cancer or the adjusted PSA > 4.0 ng/ml, then these men were recommended to undergo a prostate biopsy. If these investigations were normal or biopsy was negative, men were followed up for a total of 7 years and an end of study biopsy was performed. The end of study biopsy identified additional prostate cancers among men with neither a sspicious DRE or elevated PSA and confirmed that the controls did not have prostate cancer. A total of 18,880 eligible men were randomised into the trial. Our institution received DNA samples from 2434 cases and controls from the PCPT study.
The details of the SELECT trial can found in more detail elsewhere [21]. Healthy men entered the study and were aged 55 years or over (Afro-American men were aged 50 or older), had a normal DRE and a PSA ≤ 4 ng/ml. Participants were randomised into one of four groups: Selenium alone, vitamin E alone, selenium and vitamin E or placebo. Men were followed every 6 months and suggested to have an annual DRE and PSA. Men were recommended to undergo PSA and DRE testing and prostate biopsy based on local care guidelines. Upon a diagnosis of prostate cancer, men were monitored annually.
A total of 35,553 men were randomised into the trial. Our institution received DNA samples from 4885 men, which were samples that were selected to be analysed as part of a case-cohort study.
Laboratory methods, genotyping and quality control
DNA was collected by white blood cells from participants in the PCPT and SELECT trials and extracted at the National Cancer Institute [22]. Thirty microlitres of DNA was received at The Institute of Cancer Research; 5 µl was sent for genotyping for the purpose of this analysis.
At the time of designing the genotyping assay the latest 100 SNP panel was used [23]. Supplementary Table 2 lists the 100 known prostate cancer susceptibility SNPs at that time.
The 100 SNP panel was developed by Affymetrix®, now a part of ThermoFisher Scientific. The samples were genotyped using the Affymetrix® Eureka™ Genotyping protocols. The Eureka genotyping assay is a ligation-dependent PCR reaction, which uses interrogation site bar codes contained within the ligation probes, as well as sample index bar codes added during the amplification step. Next generation sequencing libraries were created and short cycle sequence data were generated from the prepared libraries. Analysis software was used to tabulate the number of reads that contain each combination of sample, locus and allele bar code (as appropriate). The genotype was determined by in-house software using Affymetrix® Eureka™ protocols.
In order to validate this new genotyping technique two methods were used. Firstly, known genotypes of overlapping samples from the iCOGs custom array and the Affymetrix panel were contrasted [24]. Secondly the observed minor allele frequencies from the Affymetrix panel were compared with the genotypes of overlapping samples on a custom high-throughput array by case and control status [25]. The minor allele frequencies between the two techniques were highly correlated (r2 > 0.99).
Standard quality control measures were applied to remove variants with missing rates > 10% or displayed genotype frequency deviating from those expected under Hardy–Weinberg equilibrium (P < 0.05). Samples with less than 90% genotyping rate were also removed (Table 1).
Measured outcome
On entry into the PCPT and SELECT trials, phenotypic information was collected on all participants; further detailed information on the phenotypic information collected can be found in the individual trial protocols [26, 27].
Men who self-reported to be of European origin were only included in this study as the vast majority of the germline SNPs used in this analysis were discovered in GWAS from populations of European descent. FHx was defined as men who had one or more first degree relatives affected with prostate cancer. Biopsies that were positive for prostate cancer were reviewed by the local pathologist at the participating centre, and for the PCPT trial and also reviewed centrally [8, 21]. High-grade prostate cancer (non-indolent prostate cancer) was defined as a Gleason score ≥ 7. A summary of basic phenotype information for the DNA received from PCPT and SELECT can be found here (Table 2).
Statistical analysis
A polygenic risk score was calculated by summing the genotype dosage for all variants for an individual. The log-odds ratios used to weight the risk score were taken from the OncoArray Meta-analysis [28]. Two types of risk score were calculated:
Non-weighted, for patient i:\({\mathrm{risk}\,\mathrm{score}_i} = \mathop {\sum}\nolimits_1^j {G_{ij}}\)
Weighted, for patient i: \({\mathrm{weighted}\,\mathrm{risk}\,\mathrm{score}_i} = \mathop {\sum}\nolimits_1^j {\beta _j} G_{ij}\)
j = variants 1–100
βj = is the per allele log-odds ratio for the risk of prostate cancer associated with variant j
G = risk allele dosage
Within each cohort of both trials age and body mass index (BMI) were equally distributed. Logistic regression was used to test the association of case/control and Gleason score and the polygenic risk score. FHx was also used as a covariate in the analyses. The polygenic risk score was divided into quartiles for the logistic regression model and interaction tested. For each individual SNP the χ2 test was used to test the association with overall prostate cancer and sub-strata defined by Gleason grade. Statistical significance was determined at a level of P < 0.05. All analyses were performed in the statistical package R 3.2.2 [29].
Power calculations
The power was calculated using the MAF and OR from the Oncoarray paper which was the largest GWAS performed to date [28]. A significance level of α = 0.05 was used and calculated using a genetic power calculator [30]. The results can be seen in Supplementary Table 1. The power ranged for PCPT (5–96%) and SELECT (5–99%). The power was greater in the SELECT trial as there were greater number of participants the PCPT. Overall the study was underpowered however there was a correlation between those with a high power and the signal seen of the SNP in PCPT/SELECT. A multiple comparison adjustment was not performed as known variants were analysed in PCPT/SELECT. Power was not calculated by randomisation arm as the power would have been too low.
Results
The characteristics of the participants of the trial are summarised in Table 2. Of the 100 SNPs, 98 SNPs were designed successfully. Non-Europeans were removed from the analysis and summary of the quality control is shown in Table 3.
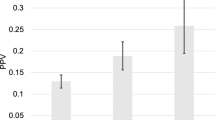
The single SNP association in both trials shows multiple SNPs that are associated with developing prostate cancer at a pre-defined significance level of P < 0.05 (Figs 1–3 and Tables 3 and 4). In the PCPT trial rs4430796 is associated in the finasteride arm with development of prostate cancer; the SNP resides near the gene HNF1B. There are also multiple SNPs which are significant for the association with high grade and low grade Gleason score (Table 5). In the PCPT trial as the polygenic risk score increases the beta predicting prostate cancer outcome increases and this is statistically significant. With the addition of Finasteride the beta reduces but it is not statistically significant (Table 6). In the SELECT trial the polygenic risk score did not predict cancer outcome except for those men who are in group 4 of the weighted risk score. There was no interaction between the interventions in the PCPT and SELECT trials and the polygenic risk score.
In the PCPT trial, the use of drug intervention did predict the outcome of developing high-grade prostate cancer, but the chemopreventions in the SELECT trial were null (Table 7). The weighted polygenic risk score was consistently higher in cases than controls in all groups and this was also statistically significant (P < 0.05) in both PCPT and SELECT trials (Figs 4 and 5). Men in the study who had a first degree relative with prostate cancer had a higher polygenic SNP score in both studies, but this was not statistically significant (P > 0.05).
Discussion
This study confirmed that men in both the PCPT and SELECT trials who developed prostate cancer had a higher polygenic risk score than men who did not develop prostate cancer. The study found no evidence that a high polygenic risk score in combination with other risk factors such as FHx could predict if the drug interventions could reduce prostate cancer incidence or development of high grade prostate cancer. Some individual SNPs were detected to predict the likelihood of developing cancer in the individual study arms but these analyses were limited by the power. Rare variants such as those found in sequencing of BRCA1 and BRCA2 were not included in this analysis as they require sequencing rather than genotyping in larger cohorts using different statistical analyses. This study was limited to prostate cancer risk associated with common variants of European ancestry and is not useable in other ethnic groups.
The results of the PCPT trial showed a 24.8% reduction in prostate cancer, but the chemoprevention in the SELECT trial dud bit reduce prostate cancer incidence [8, 9]. Prostate cancer SNPs are mostly found in intronic regions of genes, and therefore functionally it is not clear how these SNPs increase prostate cancer risk.
Genetic variations in the pathways in which the chemoprevention agents act may influence the efficacy of the agent. Finasteride acts by inhibiting the enzyme 5α reductase which is mediated by genes of the SRF5A family [31]. Polymorphisms in the SRFA genes have been reported which affect the efficacy of Finasteride [32, 33]. Genetic variations in the selenoprotein genes impact on plasma selenium levels, and recent evidence suggests that this may be associated with locally advanced or aggressive prostate cancer [14]. Variations in vitamin E levels may also be modified by genetic variations in vitamin E related genes and an association between these and a lower incidence of prostate cancer has been found [12]. If the above mechanisms that affect the outcome of these chemoprevention agents were in common functional pathways with the prostate cancer risk SNPs, men in the highest polygenic risk score would be more likely to see some of the above benefits from chemoprevention. Many of the pathways in which these genetic variants affect drug efficacy remain unknown, which therefore could be accounting for the null results of this study.
In the single SNP association for the development of prostate cancer a number of SNPs pass the significance level. Functionally it is not clear how these associations are linked but there are potentially some interesting regions. One interesting SNP which was statistically significant in the Finasteride arm of the PCPT trial was rs4430796 which resides near the gene HNF1B [34]. This SNP is in strong linkage disequilibrium with SNP rs757210. It has been reported that inheriting the risk allele for one of these SNPs increases the likelihood of developing prostate cancer (OR = 1.22, 95% CI 1.15–1.30; P = 1.4 × 10–11), but reduces the risk of developing type 2 diabetes (OR = 0.91, 95% CI 0.88–0.93; P = 8 × 10–10) [35]. The association between the phenotypes of type 2 diabetes and prostate cancer was further investigated in the PCPT trial. This showed that type 2 diabetes was associated with a 47% reduction of low grade prostate cancer and 28% reduction of high grade prostate cancer [36]. When the authors looked at the association of obesity and prostate cancer they found that increased obesity reduced low grade prostate cancer but increased high grade prostate cancer [36].The authors also showed that there was no correlation between obesity and type 2 diabetes, suggesting an independent pathway in which diabetes protects against prostate cancer. Our analysis supports these data which shows that diabetes incidence is lower in men who develop prostate cancer in the placebo arm and inherit one of the SNPs near HNF1B. However this association is not seen in men who have Finasteride who have higher rates of type 2 diabetes and could possibly suggest a metabolic interaction between the drug and the SNP.
On analyses of the SNP association with high grade and low grade Gleason score a number of SNPs show an association with Gleason grade in the individual trial arms. An example in the Finasteride arm of the PCPT, SNP rs7127900 showed a statistically significant reduction in the odds ratio of developing high grade prostate cancer (P = 0.04, OR = 0.67). Interestingly SNP rs7127900 has been shown to have a biological interaction with insulin-like-growth-factor-2 [37]. Analyses from the PCPT trial have shown that serum levels of IGF were not correlated with prostate cancer development; however, men who have high levels of IGF are more likely to be on anti-diabetic drugs such as metformin which have shown to have anti-cancer properties [38]. In the SELECT trial SNP rs12621278 in the vitamin E arm was also associated with a reduced odds ratio of developing high grade prostate cancer. The SNP rs12621278 resides near the integrin gene ITGA6 which has been shown to be associated with prostate cancer progression and development [39]. Vitamin E could possibly be interacting with this pathway [40].
So far 33% of common genetic variants that predict the familial risk of prostate cancer have been discovered [16]. Men in the top 1% risk distribution have a 5.7-fold relative risk of developing prostate cancer compared with the average population being profiled [24]. The National Institutes of Health GAME-ON initiative has discovered a further 63 new prostate cancer susceptibility SNPs. Further work needs to be performed to understand if these new SNPs will help understand the biological rationale for chemoprevention [28].
In summary this work has shown that a high polygenic risk score can predict the development of prostate cancer but there is no interaction with chemoprevention agents such as finasteride and selenium/vitamin E. This is an important null finding as population risk stratification will be undertaken in coming years for disease detection and prevention strategies. There is therefore no evidence from these results that certain risk groups are individually more likely to benefit from these two types of chemoprevention and other types of agent will need to be tested to try to reduce risk of high grade cancers in men with higher polygenic risk scores.
References
Globocan. Prostate cancer: estimated incidence, mortality and prevalence worldwide in 2012. 2012. http://globocan.iarc.fr/old/FactSheets/cancers/prostate-new.asp.
Eckersberger E, Finkelstein J, Sadri H, Margreiter M, Taneja SS, Lepor H, et al. Screening for prostate cancer: a review of the ERSPC and PLCO trials. Rev Urol. 2009;11:127–33.
Epstein JI, Herawi M. Prostate needle biopsies containing prostatic intraepithelial neoplasia or atypical foci suspicious for carcinoma: implications for patient care. J Urol. 2006;175:820–34.
Sandhu GS, Nepple KG, Tanagho YS, Andriole GL. Prostate cancer chemoprevention. Semin Oncol. 2013;40:276–85.
Lee J, Demissie K, Lu SE, Rhoads GG. Cancer incidence among Korean-American immigrants in the United States and native Koreans in South Korea. Cancer Control: J Moffitt Cancer Cent 2007;14:78–85.
Bosland MC. Is there a future for chemoprevention of prostate cancer? Cancer Prev Res. 2016;9:642–7. https://doi.org/10.1158/1940-6207.
Higgins B, Thompson IM. The prostate cancer prevention trial: current status. J Urol. 2004;171:S15–7. discussion S8.
Thompson IM, Goodman PJ, Tangen CM, Lucia MS, Miller GJ, Ford LG, et al. The influence of finasteride on the development of prostate cancer. N Engl J Med. 2003;349:215–24.
Klein EA, Thompson IM Jr., Tangen CM, Crowley JJ, Lucia MS, Goodman PJ, et al. Vitamin E and the risk of prostate cancer: the selenium and vitamin E cancer prevention trial (SELECT). J Am Med Assoc. 2011;306:1549–56.
Thompson IM Jr., Goodman PJ, Tangen CM, Parnes HL, Minasian LM, Godley PA, et al. Long-term survival of participants in the prostate cancer prevention trial. N Engl J Med. 2013;369:603–10.
Chan JM, Darke AK, Penney KL, Tangen CM, Goodman PJ, Lee GS, et al. Selenium- or vitamin E-related gene variants, interaction with supplementation, and risk of high-grade prostate cancer in SELECT. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive. Oncology. 2016;25:1050–8.
Major JM, Yu K, Weinstein SJ, Berndt SI, Hyland PL, Yeager M, et al. Genetic variants reflecting higher vitamin e status in men are associated with reduced risk of prostate cancer. J Nutr. 2014;144:729–33.
Tang L, Yao S, Till C, Goodman PJ, Tangen CM, Wu Y, et al. Repeat polymorphisms in estrogen metabolism genes and prostate cancer risk: results from the Prostate Cancer Prevention Trial. Carcinogenesis 2011;32:1500–6.
Xie W, Yang M, Chan J, Sun T, Mucci LA, Penney KL, et al. Association of genetic variations of selenoprotein genes, plasma selenium levels, and prostate cancer aggressiveness at diagnosis. Prostate. 2016;76:691–9.
Eeles R, Goh C, Castro E, Bancroft E, Guy M, Al Olama AA, et al. The genetic epidemiology of prostate cancer and its clinical implications. Nat Rev Urol. 2014;11:18–31.
Al Olama AA, Kote-Jarai Z, Berndt SI, Conti DV, Schumacher F, Han Y, et al. A meta-analysis of 87,040 individuals identifies 23 new susceptibility loci for prostate cancer. Nat Genet. 2014;46:1103–9.
Tsuchihashi Z, Dracopoli NC. Progress in high throughput SNP genotyping methods. Pharmacogenomics J. 2002;2:103–10.
Nielsen R, Paul JS, Albrechtsen A, Song YS. Genotype and SNP calling from next-generation sequencing data. Nat Rev Genet. 2011;12:443–51.
Hazelett DJ, Rhie SK, Gaddis M, Yan C, Lakeland DL, Coetzee SG, et al. Comprehensive functional annotation of 77 prostate cancer risk loci. PLoS Genet. 2014;10:e1004102.
Phillips KA, Milne RL, Rookus MA, Daly MB, Antoniou AC, Peock S, et al. Tamoxifen and risk of contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. J Clin Oncol. 2013;31:3091–9.
Lippman SM, Klein EA, Goodman PJ, Lucia MS, Thompson IM, Ford LG, et al. Effect of selenium and vitamin E on risk of prostate cancer and other cancers: the Selenium and Vitamin E Cancer Prevention Trial (SELECT). J Am Med Assoc. 2009;301:39–51.
Hoque A, Goodman P, Ambrosone CB, Figg WD, Price DK, Kopp W, et al. Extraction of DNA from serum for high-throughput genotyping: findings from pilot studies within the Prostate Cancer Prevention Trial. Urology. 2008;71:967–70.
Ahmed M, Eeles RA. Germline genetic profiling in prostate cancer: latest developments and potential clinical applications. Future Science OA. 2016;2:FSO87. https://doi.org/10.4155/fso.15.87.
Eeles RA, Olama AA, Benlloch S, Saunders EJ, Leongamornlert DA, Tymrakiewicz M, et al. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nature Genet. 2013;45:385–91, 91e1-2.
Amos CI, Dennis J, Wang Z, Byun J, Schumacher FR, Gayther SA, et al. The OncoArray Consortium: A Network for Understanding the Genetic Architecture of Common Cancers. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive. Oncology. 2017;26:126–35.
Group SWO. PCPT Resources Online: South Western Oncology Group. http://swog.org/Visitors/pcpt/.
Group SWO. SELECT resources: South Western Oncology Group. http://swog.org/Visitors/select/.
Schumacher FR, Al Olama AA, Berndt SI, Benlloch S, Ahmed M, Saunders EJ, et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat Genet. 2018;50:928–36.
Team RC. R: A language and environment for statistical computing. Austria: R Foundation for Statistical Computing Vienna; 2014. http://www.R-project.org/.
Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–50.
Price DK, Chau CH, Till C, Goodman PJ, Leach RJ, Johnson-Pais TL, et al. Association of androgen metabolism gene polymorphisms with prostate cancer risk and androgen concentrations: Results from the Prostate Cancer Prevention Trial. Cancer. 2016;122:2332–40. https://doi.org/10.1002/cncr.30071.
Makridakis NM, di Salle E, Reichardt JK. Biochemical and pharmacogenetic dissection of human steroid 5 alpha-reductase type II. Pharmacogenetics.2000;10:407–13.
Makridakis N, Akalu A, Reichardt JK. Identification and characterization of somatic steroid 5alpha-reductase (SRD5A2) mutations in human prostate cancer tissue. Oncogene 2004;23:7399–405.
Painter JN, O’Mara TA, Batra J, Cheng T, Lose FA, Dennis J, et al. Fine-mapping of the HNF1B multicancer locus identifies candidate variants that mediate endometrial cancer risk. Hum Mol Genet. 2015;24:1478–92.
Frayling TM, Colhoun H, Florez JC. A genetic link between type 2 diabetes and prostate cancer. Diabetologia. 2008;51:1757–60.
Gong Z, Neuhouser ML, Goodman PJ, Albanes D, Chi C, Hsing AW, et al. Obesity, diabetes, and risk of prostate cancer: results from the prostate cancer prevention trial. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive. Oncology. 2006;15:1977–83.
Tao S, Wang Z, Feng J, Hsu FC, Jin G, Kim ST, et al. A genome-wide search for loci interacting with known prostate cancer risk-associated genetic variants. Carcinogenesis. 2012;33:598–603.
Chae YK, Arya A, Malecek MK, Shin DS, Carneiro B, Chandra S, et al. Repurposing metformin for cancer treatment: current clinical studies. Oncotarget. 2016;7:40767–80. https://doi.org/10.18632/oncotarget.8194.
Cheng I, Plummer SJ, Neslund-Dudas C, Klein EA, Casey G, Rybicki BA, et al. Prostate cancer susceptibility variants confer increased risk of disease progression. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive. Oncology. 2010;19:2124–32.
Bureyko T, Hurdle H, Metcalfe JB, Clandinin MT, Mazurak VC. Reduced growth and integrin expression of prostate cells cultured with lycopene, vitamin E and fish oil in vitro. Br J Nutr. 2009;101:990–7.
Funding
Genotyping of the OncoArray was funded by the US National Institutes of Health (NIH) [U19 CA 148537 for ELucidating Loci Involved in Prostate cancer SuscEptibility (ELLIPSE) project and X01HG007492 to the Center for Inherited Disease Research (CIDR) under contract number HHSN268201200008I]. Additional analytic support was provided by NIH NCI U01 CA188392 (PI: Schumacher). The PRACTICAL consortium was supported by Cancer Research UK Grants C5047/A7357, C1287/A10118, C1287/A16563, C5047/A3354, C5047/A10692, C16913/A6135, European Commission’s Seventh Framework Programme grant agreement no 223175 (HEALTH-F2-2009-223175), and The National Institute of Health (NIH) Cancer Post-Cancer GWAS initiative grant: No. 1 U19 CA 148537-01 (the GAME-ON initiative). We would also like to thank the following for funding support: The Institute of Cancer Research and The Everyman Campaign, The Prostate Cancer Research Foundation, Prostate Research Campaign UK (now Prostate Action), The Orchid Cancer Appeal, The National Cancer Research Network UK, The National Cancer Research Institute (NCRI) UK. We are grateful for support of NIHR funding to the NIHR Biomedical Research Centre at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust. Research reported in this publication was supported in part by the National Cancer Institute of the National Institutes of Health under Award Numbers U10 CA37429 and UM1CA182883. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Thanks to the study leadership of PCPT and SELECT for sharing their samples and clinical data.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical considerations
All studies have the necessary ethical approval and documentation can be provided if necessary
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Members from the Prostate Cancer Association Group to Investigate Cancer Associated Alterations in the Genome (PRACTICAL) consortium are provided in the Supplement/foot notes. Information of the consortium can be found at http://practical.icr.ac.uk/.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ahmed, M., Goh, C., Saunders, E. et al. Germline genetic variation in prostate susceptibility does not predict outcomes in the chemoprevention trials PCPT and SELECT. Prostate Cancer Prostatic Dis 23, 333–342 (2020). https://doi.org/10.1038/s41391-019-0181-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41391-019-0181-y
This article is cited by
-
Inherited risk assessment and its clinical utility for predicting prostate cancer from diagnostic prostate biopsies
Prostate Cancer and Prostatic Diseases (2022)