Abstract
Pathogen-associated molecular patterns (PAMPs) have the capacity to couple inflammatory gene expression to changes in macrophage metabolism, both of which influence subsequent inflammatory activities. Similar to their microbial counterparts, several self-encoded damage-associated molecular patterns (DAMPs) induce inflammatory gene expression. However, whether this symmetry in host responses between PAMPs and DAMPs extends to metabolic shifts is unclear. Here, we report that the self-encoded oxidized phospholipid oxPAPC alters the metabolism of macrophages exposed to lipopolysaccharide. While cells activated by lipopolysaccharide rely exclusively on glycolysis, macrophages exposed to oxPAPC also use mitochondrial respiration, feed the Krebs cycle with glutamine, and favor the accumulation of oxaloacetate in the cytoplasm. This metabolite potentiates interleukin-1β production, resulting in hyperinflammation. Similar metabolic adaptions occur in vivo in hypercholesterolemic mice and human subjects. Drugs that interfere with oxPAPC-driven metabolic changes reduce atherosclerotic plaque formation in mice, thereby underscoring the importance of DAMP-mediated activities in pathophysiological conditions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon request. Uncropped raw immunoblot images can be found in the ‘Source data’ section of the Supplementary Information. Participant-level phenotype and genotype data from the FHS are accessible from the US National Center for Biotechnology Information database of Genotypes and Phenotypes at https://dbgap.ncbi.nlm.nih.gov/ to approved scientific investigators pursuing research questions that are consistent with the informed consent agreements provided by individual research participants. The FHS expression data are available from the database of Genotypes and Phenotypes at https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000363.v3.p6.
References
Brubaker, S. W., Bonham, K. S., Zanoni, I. & Kagan, J. C. Innate immune pattern recognition: a cell biological perspective. Annu. Rev. Immunol. 33, 257–290 (2015).
Janeway, C. A. Jr Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 54, 1–13 (1989).
Matzinger, P. Tolerance, danger, and the extended family. Annu. Rev. Immunol. 12, 991–1045 (1994).
Nathan, C. Points of control in inflammation. Nature 420, 846–852 (2002).
Iwasaki, A. & Medzhitov, R. Regulation of adaptive immunity by the innate immune system. Science 327, 291–295 (2010).
Zanoni, I., Tan, Y., Di Gioia, M., Springstead, J. R. & Kagan, J. C. By capturing inflammatory lipids released from dying cells, the receptor CD14 induces inflammasome-dependent phagocyte hyperactivation. Immunity 47, 697–709 (2017).
Zanoni, I. et al. An endogenous caspase-11 ligand elicits interleukin-1 release from living dendritic cells. Science 352, 1232–1236 (2016).
Imai, Y. et al. Identification of oxidative stress and Toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell 133, 235–249 (2008).
Shirey, K. A. et al. The TLR4 antagonist Eritoran protects mice from lethal influenza infection. Nature 497, 498–502 (2013).
Berliner, J. A., Leitinger, N. & Tsimikas, S. The role of oxidized phospholipids in atherosclerosis. J. Lipid Res. 50, S207–S212 (2009).
Chang, M. K. et al. Apoptotic cells with oxidation-specific epitopes are immunogenic and proinflammatory. J. Exp. Med. 200, 1359–1370 (2004).
Bochkov, V. N. et al. Protective role of phospholipid oxidation products in endotoxin-induced tissue damage. Nature 419, 77–81 (2002).
Leitinger, N. Oxidized phospholipids as modulators of inflammation in atherosclerosis. Curr. Opin. Lipidol. 14, 421–430 (2003).
Que, X. et al. Oxidized phospholipids are proinflammatory and proatherogenic in hypercholesterolaemic mice. Nature 558, 301–306 (2018).
Dinarello, C. A. Interleukin-1 in the pathogenesis and treatment of inflammatory diseases. Blood 117, 3720–3732 (2011).
Jung, J., Zeng, H. & Horng, T. Metabolism as a guiding force for immunity. Nat. Cell Biol. 21, 85–93 (2019).
O’Neill, L. A. & Pearce, E. J. Immunometabolism governs dendritic cell and macrophage function. J. Exp. Med. 213, 15–23 (2016).
Jha, A. K. et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 42, 419–430 (2015).
Van den Bossche, J. et al. Mitochondrial dysfunction prevents repolarization of inflammatory macrophages. Cell Rep. 17, 684–696 (2016).
Serbulea, V. et al. Macrophage phenotype and bioenergetics are controlled by oxidized phospholipids identified in lean and obese adipose tissue. Proc. Natl Acad. Sci. USA 115, E6254–E6263 (2018).
Everts, B. et al. TLR-driven early glycolytic reprogramming via the kinases TBK1-IKKɛ supports the anabolic demands of dendritic cell activation. Nat. Immunol. 15, 323–332 (2014).
Tannahill, G. M. et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 496, 238–242 (2013).
Palsson-McDermott, E. M. et al. Pyruvate kinase M2 regulates Hif-1α activity and IL-1β induction and is a critical determinant of the Warburg effect in LPS-activated macrophages. Cell Metab. 21, 65–80 (2015).
Serbulea, V. et al. Macrophages sensing oxidized DAMPs reprogram their metabolism to support redox homeostasis and inflammation through a TLR2-Syk-ceramide dependent mechanism. Mol. Metab. 7, 23–34 (2018).
Mills, E. L. et al. Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell 167, 457–470 (2016).
Bailey, J. D. et al. Nitric oxide modulates metabolic remodeling in inflammatory macrophages through TCA cycle regulation and itaconate accumulation. Cell Rep. 28, 218–230 (2019).
Meiser, J. et al. Pro-inflammatory macrophages sustain pyruvate oxidation through pyruvate dehydrogenase for the synthesis of itaconate and to enable cytokine expression. J. Biol. Chem. 291, 3932–3946 (2016).
Wang, F. et al. Glycolytic stimulation is not a requirement for M2 macrophage differentiation. Cell Metab. 28, 463–475 (2018).
Liu, P. S. et al. α-Ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat. Immunol. 18, 985–994 (2017).
Lampropoulou, V. et al. Itaconate links inhibition of succinate dehydrogenase with macrophage metabolic remodeling and regulation of inflammation. Cell Metab. 24, 158–166 (2016).
Koivunen, P. et al. Inhibition of hypoxia-inducible factor (HIF) hydroxylases by citric acid cycle intermediates: possible links between cell metabolism and stabilization of HIF. J. Biol. Chem. 282, 4524–4532 (2007).
Ridker, P. M. et al. Antiinflammatory therapy with canakinumab for atherosclerotic disease. New Engl. J. Med. 377, 1119–1131 (2017).
Koelwyn, G. J., Corr, E. M., Erbay, E. & Moore, K. J. Regulation of macrophage immunometabolism in atherosclerosis. Nat. Immunol. 19, 526–537 (2018).
Steinberg, D. & Witztum, J. L. Oxidized low-density lipoprotein and atherosclerosis. Arter. Thromb. Vasc. Biol. 30, 2311–2316 (2010).
Oskolkova, O. V. et al. Oxidized phospholipids are more potent antagonists of lipopolysaccharide than inducers of inflammation. J. Immunol. 185, 7706–7712 (2010).
Kim, K. et al. Transcriptome analysis reveals nonfoamy rather than foamy plaque macrophages are proinflammatory in atherosclerotic murine models. Circ. Res. 123, 1127–1142 (2018).
Cochain, C. et al. Single-cell RNA-Seq reveals the transcriptional landscape and heterogeneity of aortic macrophages in murine atherosclerosis. Circ. Res. 122, 1661–1674 (2018).
Mahmood, S. S., Levy, D., Vasan, R. S. & Wang, T. J. The Framingham Heart Study and the epidemiology of cardiovascular disease: a historical perspective. Lancet 383, 999–1008 (2014).
Sanin, D. E. et al. Mitochondrial membrane potential regulates nuclear gene expression in macrophages exposed to prostaglandin E2. Immunity 49, 1021–1033 (2018).
Johnson, M. O. et al. Distinct regulation of Th17 and Th1 cell differentiation by glutaminase-dependent metabolism. Cell 175, 1780–1795 (2018).
Shirai, T. et al. The glycolytic enzyme PKM2 bridges metabolic and inflammatory dysfunction in coronary artery disease. J. Exp. Med. 213, 337–354 (2016).
Tavakoli, S. et al. Characterization of macrophage polarization states using combined measurement of 2-deoxyglucose and glutamine accumulation: implications for imaging of atherosclerosis. Arter. Thromb. Vasc. Biol. 37, 1840–1848 (2017).
Hitzel, J. et al. Oxidized phospholipids regulate amino acid metabolism through MTHFD2 to facilitate nucleotide release in endothelial cells. Nat. Commun. 9, 2292 (2018).
Bekkering, S. et al. Metabolic induction of trained immunity through the mevalonate pathway. Cell 172, 135–146 (2018).
Christ, A. et al. Western diet triggers NLRP3-dependent innate immune reprogramming. Cell 172, 162–175 (2018).
Geng, S. et al. The persistence of low-grade inflammatory monocytes contributes to aggravated atherosclerosis. Nat. Commun. 7, 13436 (2016).
Michelsen, K. S. et al. Lack of Toll-like receptor 4 or myeloid differentiation factor 88 reduces atherosclerosis and alters plaque phenotype in mice deficient in apolipoprotein E. Proc. Natl Acad. Sci. USA 101, 10679–10684 (2004).
Carnevale, R. et al. Localization of lipopolysaccharide from Escherichia coli into human atherosclerotic plaque. Sci. Rep. 8, 3598 (2018).
Philippova, M. et al. Analysis of fragmented oxidized phosphatidylcholines in human plasma using mass spectrometry: comparison with immune assays. Free Radic. Biol. Med. 144, 167–175 (2019).
Bjorkbacka, H. et al. Reduced atherosclerosis in MyD88-null mice links elevated serum cholesterol levels to activation of innate immunity signaling pathways. Nat. Med. 10, 416–421 (2004).
Yuan, M., Breitkopf, S. B., Yang, X. & Asara, J. M. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat. Protoc. 7, 872–881 (2012).
Watson, A. D. et al. Structural identification of a novel pro-inflammatory epoxyisoprostane phospholipid in mildly oxidized low density lipoprotein. J. Biol. Chem. 274, 24787–24798 (1999).
Yuan, M. et al. Ex vivo and in vivo stable isotope labelling of central carbon metabolism and related pathways with analysis by LC-MS/MS. Nat. Protoc. 14, 313–330 (2019).
Kannel, W. B., Feinleib, M., McNamara, P. M., Garrison, R. J. & Castelli, W. P. An investigation of coronary heart disease in families. The Framingham Offspring Study. Am. J. Epidemiol. 110, 281–290 (1979).
Joehanes, R. et al. Gene expression signatures of coronary heart disease. Arter. Thromb. Vasc. Biol. 33, 1418–1426 (2013).
Irizarry, R. A. et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 (2003).
Joehanes, R. et al. Integrated genome-wide analysis of expression quantitative trait loci aids interpretation of genomic association studies. Genome Biol. 18, 16 (2017).
Acknowledgements
We thank F. Granucci, J. C. Kagan and L. R. Marek for discussions, help and support. R.S. thanks the UCLA QCBio Collaboratory community, directed by M. Pellegrini. I.Z. is supported by National Institutes of Health (NIH) grants 1R01AI121066, 1R01DK115217 and NIAID-DAIT-NIHAI201700100. J.R.S. is supported by NIH grant 1R15HL121770-01A1. The FHS is funded by NIH contracts N01-HC-25195 and HHSN268201500001I. The laboratory work for the FHS investigation was funded by the Division of Intramural Research, National Heart, Lung, and Blood Institute, NIH, and by a Director’s Challenge Award, NIH (principal investigator: D.L.). This study utilized the computational resources of the Biowulf system at the NIH in Bethesda, Maryland (http://hpc.nih.gov). M.M.M. is supported by NIH grant K99HL136875. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Author information
Authors and Affiliations
Contributions
M.D.G. designed, performed and analyzed the experiments. R.S. performed the statistical comparisons on the FHS aggregated data. J.R.S. produced oxPAPC and PEIPC, and participated in the analyses of data. M.M.M., R.J. and D.L. developed and analyzed the FHS expression project. I.Z. conceived the project, designed the experiments, supervised the study and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
I.Z., M.D.G. and Boston Children’s Hospital have filed an international patent application (US patent application number 62/851,561) that relates to the metabolic activity of oxPAPC.
Additional information
Peer review information Zoltan Fehervari was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
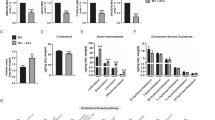
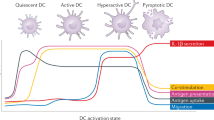
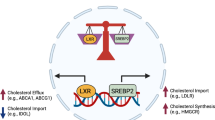
Extended Data Fig. 1 oxPAPC drives hyperactivation and hypermetabolism in macrophages.
Schematic depicting oxPAPC activities. oxPAPC is a mixture of oxidized phospholipids that induce an hyperinflammatory state in phagocytes upon LPS encounter and/or during atherosclerosis development. Moieties such as POVPC or PGPC contained in oxPAPC drive the formation of hyperactive cells that are characterized by inflammasome activation in the absence of pyroptosis. In contrast to POVPC or PGPC, PEIPC engages a hypermetabolic state in phagocytes that favors IL-1β accumulation and that is characterized by: i) the simultaneous activation of OXPHOS and aerobic glycolysis; ii) glutamine utilization to feed the TCA cycle; iii) oxaloacetate (OAA) accumulation in the cytoplasm to potentiate HIF-1α activation.
Supplementary information
Supplementary Information
Supplementary Figs. 1–8.
Supplementary Table 1
Metabolomic analysis of BMDMs primed for 3 h with LPS (1 μg ml−1) and treated, or not, with oxPAPC (100 μg ml−1).
Supplementary Table 2
FHS gene expression analysis.
Source data
Source Data Fig. 1
Statistical Data
Source Data Fig. 2
Statistical Data
Source Data Fig. 3
Statistical Data
Source Data Fig. 4
Statistical Data
Source Data Fig. 5
Statistical Data
Source Data Fig. 6
Statistical Data
Source Data Fig. 7
Statistical Data
Source Data Fig. 8
Statistical Data
Source Data, Supplementary Fig. 1
Statistical Data
Source Data, Supplementary Fig. 2
Statistical Data
Source Data, Supplementary Fig. 3
Statistical Data
Source Data, Supplementary Fig. 4
Statistical Data
Source Data, Supplementary Fig. 5
Statistical Data
Source Data, Supplementary Fig. 6
Statistical Data
Source Data, Supplementary Fig. 7
Statistical Data
Source Data, Supplementary Fig. 8
Statistical Data
Source Data Fig. 3
Unprocessed WB
Source Data Fig. 5
Unprocessed WB
Source Data Fig. 6
Unprocessed WB
Source Data Supplementary Fig. 3
Unprocessed WB
Source Data Supplementary Fig. 6
Unprocessed WB
Rights and permissions
About this article
Cite this article
Di Gioia, M., Spreafico, R., Springstead, J.R. et al. Endogenous oxidized phospholipids reprogram cellular metabolism and boost hyperinflammation. Nat Immunol 21, 42–53 (2020). https://doi.org/10.1038/s41590-019-0539-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-019-0539-2
This article is cited by
-
Anti-inflammatory and anti-oxidative electrospun nanofiber membrane promotes diabetic wound healing via macrophage modulation
Journal of Nanobiotechnology (2024)
-
Peroxiredoxin 3 has a crucial role in the macrophage polarization by regulating mitochondrial homeostasis
Respiratory Research (2024)
-
Dysregulated cellular metabolism in atherosclerosis: mediators and therapeutic opportunities
Nature Metabolism (2024)
-
Profiling of microglia nodules in multiple sclerosis reveals propensity for lesion formation
Nature Communications (2024)
-
Metabolism, metabolites, and macrophages in cancer
Journal of Hematology & Oncology (2023)