Abstract
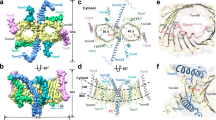
Nearly all mitochondrial proteins are encoded by the nuclear genome and imported into mitochondria after synthesis on cytosolic ribosomes. These precursor proteins are translocated into mitochondria by the TOM complex, a protein-conducting channel in the mitochondrial outer membrane. We have determined high-resolution cryo-EM structures of the core TOM complex from Saccharomyces cerevisiae in dimeric and tetrameric forms. Dimeric TOM consists of two copies each of five proteins arranged in two-fold symmetry: pore-forming β-barrel protein Tom40 and four auxiliary α-helical transmembrane proteins. The pore of each Tom40 has an overall negatively charged inner surface attributed to multiple functionally important acidic patches. The tetrameric complex is essentially a dimer of dimeric TOM, which may be capable of forming higher-order oligomers. Our study reveals the detailed molecular organization of the TOM complex and provides new insights about the mechanism of protein translocation into mitochondria.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Wiedemann, N. & Pfanner, N. Mitochondrial machineries for protein import and assembly. Annu. Rev. Biochem. 86, 685–714 (2017).
Chacinska, A., Koehler, C. M., Milenkovic, D., Lithgow, T. & Pfanner, N. Importing mitochondrial proteins: machineries and mechanisms. Cell 138, 628–644 (2009).
Neupert, W. & Herrmann, J. M. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76, 723–749 (2007).
Kiebler, M. et al. Identification of a mitochondrial receptor complex required for recognition and membrane insertion of precursor proteins. Nature 348, 610–616 (1990).
Kunkele, K. P. et al. The preprotein translocation channel of the outer membrane of mitochondria. Cell 93, 1009–1019 (1998).
Dekker, P. J. et al. Preprotein translocase of the outer mitochondrial membrane: molecular dissection and assembly of the general import pore complex. Mol. Cell Biol. 18, 6515–6524 (1998).
Ahting, U. et al. The TOM core complex: the general protein import pore of the outer membrane of mitochondria. J. Cell Biol. 147, 959–968 (1999).
Rapaport, D. et al. Dynamics of the TOM complex of mitochondria during binding and translocation of preproteins. Mol. Cell Biol. 18, 5256–5262 (1998).
Meisinger, C. et al. Protein import channel of the outer mitochondrial membrane: a highly stable Tom40-Tom22 core structure differentially interacts with preproteins, small Tom proteins, and import receptors. Mol. Cell Biol. 21, 2337–2348 (2001).
Schmitt, S. et al. Role of Tom5 in maintaining the structural stability of the TOM complex of mitochondria. J. Biol. Chem. 280, 14499–14506 (2005).
Hill, K. et al. Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins. Nature 395, 516–521 (1998); comment 395, 439−440.
Ahting, U. et al. Tom40, the pore-forming component of the protein-conducting TOM channel in the outer membrane of mitochondria. J. Cell Biol. 153, 1151–1160 (2001).
Shiota, T. et al. Molecular architecture of the active mitochondrial protein gate. Science 349, 1544–1548 (2015).
Zeth, K. Structure and evolution of mitochondrial outer membrane proteins of beta-barrel topology. Biochim. Biophys. Acta 1797, 1292–1299 (2010).
Lackey, S. W. et al. Evidence supporting the 19 beta-strand model for Tom40 from cysteine scanning and protease site accessibility studies. J. Biol. Chem. 289, 21640–21650 (2014).
Bolliger, L., Junne, T., Schatz, G. & Lithgow, T. Acidic receptor domains on both sides of the outer membrane mediate translocation of precursor proteins into yeast mitochondria. EMBO J. 14, 6318–6326 (1995).
Dietmeier, K. et al. Tom5 functionally links mitochondrial preprotein receptors to the general import pore. Nature 388, 195–200 (1997).
Abe, Y. et al. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell 100, 551–560 (2000).
Yamano, K. et al. Tom20 and Tom22 share the common signal recognition pathway in mitochondrial protein import. J. Biol. Chem. 283, 3799–3807 (2008).
Yamamoto, H. et al. Roles of Tom70 in import of presequence-containing mitochondrial proteins. J. Biol. Chem. 284, 31635–31646 (2009).
Qiu, J. et al. Coupling of mitochondrial import and export translocases by receptor-mediated supercomplex formation. Cell 154, 596–608 (2013).
Sherman, E. L., Go, N. E. & Nargang, F. E. Functions of the small proteins in the TOM complex of Neurospora crassa. Mol. Biol. Cell 16, 4172–4182 (2005).
Becker, T. et al. Biogenesis of mitochondria: dual role of Tom7 in modulating assembly of the preprotein translocase of the outer membrane. J. Mol. Biol. 405, 113–124 (2011).
Rapaport, D., Neupert, W. & Lill, R. Mitochondrial protein import. Tom40 plays a major role in targeting and translocation of preproteins by forming a specific binding site for the presequence. J Biol. Chem. 272, 18725–18731 (1997).
Melin, J. et al. Presequence recognition by the Tom40 channel contributes to precursor translocation into the mitochondrial matrix. Mol. Cell Biol. 34, 3473–3485 (2014).
Vogtle, F. N. et al. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell 139, 428–439 (2009).
Bausewein, T. et al. Cryo-EM Structure of the TOM core complex from Neurospora crassa. Cell 170, 693–700.e7 (2017).
Model, K. et al. Protein translocase of the outer mitochondrial membrane: role of import receptors in the structural organization of the TOM complex. J. Mol. Biol. 316, 657–666 (2002).
Model, K., Meisinger, C. & Kuhlbrandt, W. Cryo-electron microscopy structure of a yeast mitochondrial preprotein translocase. J. Mol. Biol. 383, 1049–1057 (2008).
Hauer, F. et al. GraDeR: Membrane protein complex preparation for single-particle cryo-EM. Structure 23, 1769–1775 (2015).
Ujwal, R. et al. The crystal structure of mouse VDAC1 at 2.3 Å resolution reveals mechanistic insights into metabolite gating. Proc. Natl Acad. Sci. USA 105, 17742–17747 (2008).
Allen, R., Egan, B., Gabriel, K., Beilharz, T. & Lithgow, T. A conserved proline residue is present in the transmembrane-spanning domain of Tom7 and other tail-anchored protein subunits of the TOM translocase. FEBS Lett. 514, 347–350 (2002).
Moczko, M. et al. The intermembrane space domain of mitochondrial Tom22 functions as a trans binding site for preproteins with N-terminal targeting sequences. Mol. Cell Biol. 17, 6574–6584 (1997).
Albrecht, R. et al. The Tim21 binding domain connects the preprotein translocases of both mitochondrial membranes. EMBO Rep. 7, 1233–1238 (2006).
van Wilpe, S. et al. Tom22 is a multifunctional organizer of the mitochondrial preprotein translocase. Nature 401, 485–489 (1999).
Shiota, T., Mabuchi, H., Tanaka-Yamano, S., Yamano, K. & Endo, T. In vivo protein-interaction mapping of a mitochondrial translocator protein Tom22 at work. Proc. Natl Acad. Sci. USA 108, 15179–15183 (2011).
Brix, J., Rudiger, S., Bukau, B., Schneider-Mergener, J. & Pfanner, N. Distribution of binding sequences for the mitochondrial import receptors Tom20, Tom22, and Tom70 in a presequence-carrying preprotein and a non-cleavable preprotein. J. Biol. Chem. 274, 16522–16530 (1999).
Sherman, E. L., Taylor, R. D., Go, N. E. & Nargang, F. E. Effect of mutations in Tom40 on stability of the translocase of the outer mitochondrial membrane (TOM) complex, assembly of Tom40, and import of mitochondrial preproteins. J. Biol. Chem. 281, 22554–22565 (2006).
Gabriel, K., Egan, B. & Lithgow, T. Tom40, the import channel of the mitochondrial outer membrane, plays an active role in sorting imported proteins. EMBO J. 22, 2380–2386 (2003).
Honlinger, A. et al. Tom7 modulates the dynamics of the mitochondrial outer membrane translocase and plays a pathway-related role in protein import. EMBO J. 15, 2125–2137 (1996).
Bakelar, J., Buchanan, S. K. & Noinaj, N. The structure of the beta-barrel assembly machinery complex. Science 351, 180–186 (2016).
Gu, Y. et al. Structural basis of outer membrane protein insertion by the BAM complex. Nature 531, 64–69 (2016).
Hohr, A. I. C. et al. Membrane protein insertion through a mitochondrial β-barrel gate. Science 359, eaah6834 (2018).
Kunkele, K. P. et al. The isolated complex of the translocase of the outer membrane of mitochondria. Characterization of the cation-selective and voltage-gated preprotein-conducting pore. J. Biol. Chem. 273, 31032–31039 (1998).
Da Cruz, S. et al. Proteomic analysis of the mouse liver mitochondrial inner membrane. J. Biol. Chem. 278, 41566–41571 (2003).
Sakaue, H. et al. Porin associates with Tom22 to regulate the mitochondrial protein gate assembly. Mol. Cell 73, 1044–1055.e8 (2019).
Schulz, C. et al. Tim50’s presequence receptor domain is essential for signal driven transport across the TIM23 complex. J. Cell Biol. 195, 643–656 (2011).
Williams, C. C., Jan, C. H. & Weissman, J. S. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science 346, 748–751 (2014).
Harbauer, A. B. et al. Mitochondria. Cell cycle-dependent regulation of mitochondrial preprotein translocase. Science 346, 1109–1113 (2014).
Lee, M. E., DeLoache, W. C., Cervantes, B. & Dueber, J. E. A highly characterized yeast toolkit for modular, multipart assembly. ACS Synth. Biol. 4, 975–986 (2015).
McIsaac, R. et al. Fast-acting and nearly gratuitous induction of gene expression and protein depletion in Saccharomyces cerevisiae. Mol. Biol. Cell 22, 4447–4459 (2011).
Mnaimneh, S. et al. Exploration of essential gene functions via titratable promoter alleles. Cell 118, 31–44 (2004).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Tegunov, T. C. P. Real-time cryo-EM data pre-processing with Warp. Nat. Methods 16, 1146–1152 (2019).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Dolinsky, T. J., Nielsen, J. E., McCammon, J. A. & Baker, N. A. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 32, W665–W667 (2004).
Acknowledgements
We thank D. Toso for help with electron microscope operation and J. Thorner for yeast strains and antibodies. We thank J. Thorner, J. Hurley, S. Brohawn, and S. Itskanov for critical reading of the manuscript. This work was funded by UC Berkeley (E.P. and J. T.), Vallee Scholars Program (E.P.), and NSF Graduate Research Fellowship Program (K.T.; DGE-1752814).
Author information
Authors and Affiliations
Contributions
E.P. conceived the project. K.T. and E.P. performed experiments. E.P. built the atomic models. K.T. and E.P. interpreted results and wrote the manuscript. E.P. supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Katarzyna Marcinkiewicz was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Single-particle cryo-EM analysis of the dimeric core TOM complex.
a, Summary of single-particle image analysis procedure. b, A representative motion-corrected micrograph. Scale bar, 20 nm. Right panels show magnified images of selected particles outlined with white squares. The particle image size is 209 Å (width) by 209 Å (height). c, Representative class averages from 2D classification by RELION3. The box dimensions are 297 Å (width) by 297 Å (height). Classes in red boxes are likely empty micelles and thus excluded in subsequent analysis. d, Heat map showing particle orientation distribution (produced in the final 3D reconstruction by cryoSPARC2). e, Fourier shell correlation (FSC) of two independently refined half maps. Blue line, corrected masked FSC. Solid black line, unmasked FSC.
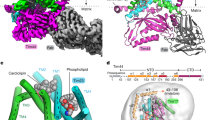
Extended Data Fig. 2 Cryo-EM map and atomic model quality of the dimeric TOM complex.
a, Local resolution represented by a heat map on the density contour (unsharpened, summed map). b, FSC between the EM map and the atomic model. Blue curve, FSCwork (FSC between half map 1 and a model refined against half map 1). Red curve, FSCfree (FSC between half map 2 and the model refined against half map 1). Black curve, FSCfull (FSC between the combined map and the final atomic model refined against the combined map. All refinements were performed by Phenix with the same weight. c, Examples of the density map and the atomic model for indicated segments. Numbers in the brackets indicate ranges of amino acid residues shown. d, Structural comparison of Tom40 and VDAC. Structures of Tom40 (this study; green) and murine VDAC (PDB 3EMN; magenta) are superimposed. Left, view from cytosol. Right, side view. e, Density features (green) for the C-terminal tails of Tom40 are shown in a 5-Å low-pass-filtered map. Shown is vertical cross-section along the Tom40 pores. A weak connection (not shown) between the density in green and α3 of Tom40 is indicated by a dashed line.
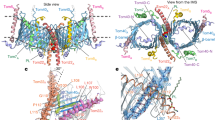
Extended Data Fig. 3 Surface complementarity of Tom subunits at interfaces and purification of the TOM complex with a K90A H102A mutation.
a, b, Overview of the dimeric TOM complex in solvent-accessible surface representation. Shown are views from the cytosol (a) and along the membrane plane (b). The color scheme is the same as in Fig. 1. The regions marked by a dashed line are magnified in c (with a 180° rotation) and d, respectively. c, Interface between the two Tom40 subunits. Left, a view from IMS (showing all subunits). Middle, as in the left panel but showing only Tom40 and DDM detergent molecules. Right, as in the middle panel but showing a side view. d,e, Side views showing the Tom40 and Tom22 interfaces within the same asymmetric unit (d) and between the two asymmetric units (e). f–h, Side view showing interfaces between Tom40 and other small Tom subunits. The viewing angles are the same as in Fig. 2b–d, respectively. i, Superose 6 SEC profile of the mutant TOM complex with K90A H102A Tom40 (solid blue) purified as the wild-type complex (dashed gray line; also see Fig. 1a).
Extended Data Fig. 4 Acidic and hydrophobic patches on the Tom40 pore surface.
a, Overview (side view) of the dimeric TOM complex (gray ribbons) and the Tom40 pore cavity (surface representation; shown for only one Tom40 subunit). b–d, Surface electrostatics is shown as a heat map overlaid on the pore cavity shown in surface representation. Side chains of acidic amino acids are shown in stick representation (AP1, AP2, and AP3 are in yellow, green, and pale blue, respectively). In c, only AP2 side chains are shown for clarity. e–g, As in b–d, but side chains of hydrophobic patches are shown in stick representation (HP1, HP2, are HP3 are in olive, green, and magenta, respectively). Note that some hydrophobic side chains in HP1 (labeled in gray; F81, F327, and L76) are only partially exposed, as they are involved in interactions between the α2 segment and the β sheets. h, As in Fig. 3j, but with mutants of AP3, HP2, and HP3. AP3mut1= E268N E308N; AP3mut2= E69N D73N; HP2mut1= L183S L216S; HP2mut2= V196N V198S; HP3mut1= L119S A121N F126N; HP3mut2= M94N A97S. Dox, doxycycline. i, Expression of Tom40 pore mutants (contains a C-terminal Strep-tag) was examined by SDS-PAGE and immunoblotting analyses of whole-cell lysates. PGK1, loading controls. Source data for i are available with the paper online. The experiments in h and i were repeated at least twice with similar results.
Extended Data Fig. 5 Homology modeled pore architecture of N. crassa Tom40.
As in Fig. 3 a–d, but with N. crassa TOM complex. An N. crassa homology model was generated by SWISS-MODEL using the S. cerevisiae structure as a template, and electrostatic potential was calculated by Adaptive Poisson-Boltzmann Solver (APBS). The dashed yellow line indicates AP2. Note that unlike the S. cerevisiae TOM complex AP3 is not prominent in N. crassa.
Extended Data Fig. 6 Effects of detergent on the oligomeric state of the TOM complex.
a, Schematic diagram of the TOM complex purification procedure. Different detergent conditions (indicated by blue texts) were tested (specific conditions in b–e). b–e, Detailed SEC profiles of the purified TOM complex purified under different detergent conditions. “D” indicates the dimer peak, and “T” indicates the tetramer peak. Positions of the void peak (void) and peaks of molecular weight standards are indicated by arrowheads. TG, thyroglobulin (670 kDa); F, ferritin (440 kDa); ald, aldolase (156 kDa). Note that b–d is the same as in Fig. 4a–c, and b is the same experiment shown in Fig. 1a. f, SDS-PAGE analysis of peak fractions from the SEC purification shown in c. The peak positions are marked with “T” and “D”. The SDS gel was stained by Coomassie. g, Crude lysates prepared from cells overexpressing the TOM complex were solubilized with indicated detergent and subjected to BN-PAGE, followed by immunoblotting using an anti-Strep-tag antibody (detecting Tom40-Strep). A gradual decrease of mobility of the TOM complex accompanied by lowered detergent concentrations is likely due to an increased detergent micelle size. Source data for g are available with the paper online. The experiment in g was repeated twice with similar results.
Extended Data Fig. 7 Cryo-EM analysis of the tetrameric complex.
a, Summary of single-particle image analysis procedure. b, A representative micrograph. Scale bar, 20 nm. The dimensions of magnified images are 414 Å (width) by 414 Å (height). c, Examples of selected 2D class averages. The box dimensions are 460 Å (width) by 460 Å (height). d, Particle orientation distribution. e, Fourier shell correlation (FSC). Blue line, corrected masked FSC. Solid black line, unmasked FSC. f, Local resolution map. g, Example images of particles larger than the tetramer. The leftmost image shows a 2D projection (side view with the longest width) of the 3D reconstruction of the tetrameric TOM complex. The other images show examples of large particles on micrographs. Estimated oligomeric states are indicated. Scale bar, 100 Å.
Extended Data Fig. 8 Dimer-dimer interface in the tetrameric TOM complex.
a, Overview (cytosolic view) of the tetrameric TOM complex. The 4.1-Å-resolution 3D reconstruction was represented with a composite map showing two different contour levels to show the protein features (colored contour; low-pass-filtered at 4.1 Å) and the detergent micelle (semitransparent gray contour; low-pass-filtered according to local resolution values). Organization of monomeric units are schematized in the upper right corner. Areas marked by dashed rectangles are shown in b and c (after rotating for a side view) with arrows and eye symbols indicating the viewing directions. b,c, Side views showing the dimer-dimer contacts between units B and C. Note that the tetramer is not symmetric and that there is a sizeable gap between Tom5B and Tom22C (c) in contrast to Tom5C and Tom22B (b). d, As in a, but showing a side view. Dashed lines indicate cross-sectional planes for cutaway views shown in e–g. e–g, Cutaway views (views from cytosol) at different positions along the membrane axis. In e and f, major interactions mediating the tetramerization are indicated by dashed ovals. Note that in g, there is a gap along the interface (also see h and i). h, As in a and d, but showing a view from IMS. i, Solvent-accessible surface of the tetrameric TOM complex. The dashed line indicates the interfacial gap. The two dimers (A–B and C–D) are in blue and red, respectively.
Extended Data Fig. 9 Biochemical validation of higher oligomeric TOM complexes.
a, Left, overview (angled cytosolic view) of the tetrameric TOM complex. Monomeric units B and C are shown in color, and A and D are in gray. The region in the black dashed box is magnified and shown in the right panel. Right, the cryo-EM density map (semitransparent gray) and the atomic model (in color) are shown for the B–C interface. The blue dashed arrows indicate the directions of the unmodeled N-terminal segments (residues 1–24) of the Tom6C and Tom6B subunits. The black dotted oval indicates the hydrophobic patch HP2. The green dashed lines indicate the unmodeled loop (L14-15; residues 277–294) between β14 and β15 of Tom40. b, Mitochondria were treated with BM-PEG2 and analyzed by SDS-PAGE and immunoblotting (IB). Tom40 contained no or an indicated single cysteine. c, Mitochondria were purified from cells expressing Tom40Strep (M287C) from the chromosomal locus and Tom40His (M287C) from a CEN plasmid. After treating with BM-PEG2, mitochondria were solubilized with octyl glucoside and subjected to immunoprecipitation (IP) using anti-Strep-tag antibodies (mock: IP without anti-Strep-tag antibodies). d, Mitochondria with Tom40Strep (M287C) expressed from the endogenous promoter were solubilized in 0.5% LMNG and 0.1% CHS and then injected to Superose 6 column. Fractions were treated with BM-PEG2 before SDS-PAGE and immunoblotting. e, As in Fig. 4e, but with mitochondria isolated from the tom6Δ mutant background. “T” and “D” indicate the peak positions of tetramers and dimers, respectively. Yeast were grown in YPEG (b and e) or YPD (c and d). Source data for panels b–e are available with the paper online. The experiments in b–e were repeated at least twice with similar results.
Supplementary information
Supplementary Table
Supplementary Table 1
Source data
Source Data Fig. 4
Uncropped Western Blots
Source Data Fig. 5
Uncropped Western Blots
Source Data Extended Data Fig. 4
Uncropped Western Blots
Source Data Extended Data Fig. 6
Uncropped Western Blots
Source Data Extended Data Fig. 9
Uncropped Western Blots
Rights and permissions
About this article
Cite this article
Tucker, K., Park, E. Cryo-EM structure of the mitochondrial protein-import channel TOM complex at near-atomic resolution. Nat Struct Mol Biol 26, 1158–1166 (2019). https://doi.org/10.1038/s41594-019-0339-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-019-0339-2
This article is cited by
-
Mitochondrial stress: a key role of neuroinflammation in stroke
Journal of Neuroinflammation (2024)
-
The architecture of substrate-engaged TOM–TIM23 supercomplex reveals preprotein proximity sites for mitochondrial protein translocation
Cell Discovery (2024)
-
Molecular pathway of mitochondrial preprotein import through the TOM–TIM23 supercomplex
Nature Structural & Molecular Biology (2023)
-
A multipoint guidance mechanism for β-barrel folding on the SAM complex
Nature Structural & Molecular Biology (2023)
-
Structural basis of mitochondrial protein import by the TIM23 complex
Nature (2023)