Abstract
PD-L1, as assessed by immunohistochemistry, is a predictive biomarker for immuno-oncology treatment in lung cancer. Different scoring methods have been used to assess its status, resulting in a wide range of positivity rates. We use the European Thoracic Oncology Platform Lungscape non-small cell lung carcinoma cohort to explore this issue. PD-L1 expression was assessed via immunohistochemistry on tissue microarrays (up to four cores per case), using the DAKO 28-8 immunohistochemistry assay, following a two-round external quality assessment procedure. All samples were analyzed under the same protocol. Cross-validation of scoring between tissue microarray and whole sections was performed in 10% randomly selected samples. Cutoff points considered: ≥1, 50 (primarily), and 25%. At the two external quality assessment rounds, tissue microarray scoring agreement rates between pathologists were: 73% and 81%. There were 2008 cases with valid immunohistochemistry tissue microarray results (50% all cores evaluable). Concordant cases at 1, 25, and 50% were: 85, 91, and 93%. Tissue microarray core results were identical for 70% of cases. Sensitivity of the tissue microarray method for 1, 25, and 50% was: 80, 78, and 79% (specificity: 90, 95, 98%). Complete agreement between tissue microarrays and whole sections was achieved for 60% of the cases. Highest sensitivity rates for 1% and 50% cutoffs were detected for higher number of cores. Underestimation of PD-L1 expression on small samples is more common than overestimation. We demonstrated that classification of PD-L1 on small biopsy samples does not represent the overall expression of PD-L1 in all non-small cell cancer carcinoma cases, although the majority of cases are ‘correctly’ classified. In future studies, sampling more and larger biopsies, recording the biopsy size and tumor load may permit further refinement, increasing predictive accuracy.
Similar content being viewed by others
Introduction
Immunohistochemical detection of the Programmed death-ligand 1 (PD-L1) protein is now an established biomarker test for the selection of patients for anti-PD1 and anti-PD-L1 immunotherapy in non-small cell lung cancer [1,2,3,4]. An introductory training in PD-L1 immunohistochemical scoring and an external quality assessment is common practice to prospectively assess alignment of pathologists scoring.
PD-L1 is often heterogeneously expressed within tumors and may differ between primary and metastatic tissues [4, 5]. It has been hypothesized that multiple tumor biopsy specimens may be needed to accurately determine the PD-L1 expression status [5]. To examine the effect of sampling on the PD-L1 score, several samples would be required from a large series of patients, however this scenario could be simulated by comparing a multicore tissue microarray with a whole tumor section.
The European Thoracic Oncology Platform established a large cohort of surgically resected non-small cell lung carcinoma cases from 15 academic centers (Lungscape). This patient cohort is clinically fully annotated and has already been extensively studied with respect to clinical outcomes and molecular analysis [6,7,8]. For PD-L1 immunohistochemistry assessment, each European Thoracic Oncology Platform Lungscape pathologist was trained in reading the assay. An external quality assessment procedure was performed by the contributing pathologists from each center.
In this paper, we first present the effect of harmonization of PD-L1 scoring on tissue microarrays, by an external quality assessment program and then the cross-validation of the tissue microarray approach vs whole section scores in European Thoracic Oncology Platform Lungscape. We examined the heterogeneity by comparing the extent of staining across different tissue microarray cores, as mimickers of individual biopsy samples, with the whole tumor, providing an impression about the accuracy of PD-L1 expression on biopsy sampling. Associations of PD-L1 and clinical and molecular features and postoperative outcomes have been presented elsewhere [9].
Materials and methods
Study design
This is a retrospective study design exploring tissue microarray core PD-L1 immunohistochemistry score variability and agreement of tissue microarray scores with whole section scores in the determination of PD-L1 status following an external quality assessment program in a cohort of surgically resected, stage I–III, non-small cell lung carcinoma cases included in the Lungscape project [6].
The study was conducted according to the Lungscape master and PD-L1 sub-study protocols; with adherence to country specific ethics, regulatory requirements, and REMARK recommendations.
External quality assessment procedure
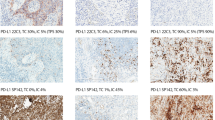
Out of the 15 European Thoracic Oncology Platform centers providing data for this project, 14 participated in the external quality assessment program. All of the 14 European Thoracic Oncology Platform pathologists received two-day face to face training for PD-L1 immunohistochemistry scoring after which there was ≥90% concordance between the DAKO trainer and European Thoracic Oncology Platform pathologists. Concordance in PD-L1 scoring between European Thoracic Oncology Platform pathologists was examined three months later via an external quality assessment procedure. To this end a tissue microarray was constructed consisting of four cell lines and eight resection specimens (two cores each, prepared in VUmc, Amsterdam, NL), representing various PD-L1 expression levels from negative to 100%. This external quality assessment tissue microarray was centrally cut and stained according to DAKO PD-L1 immunohistochemistry 28-8 pharmDx™ manufacturers protocol. Core size was similar for all participants (0.6 mm), comparable with bronchial biopsy.
Cores were photographed. Visual inspection (judged by two pathologists: KK, ET) of the aligned images did not reveal differences between the cores. The external quality assessment program was extended with 65 images of PD-L1 stained non-small cell lung carcinoma, with the request to define: the number and proportion of PD-L1 positive tumor cells. From this image set, read by 12 out of the 14 European Thoracic Oncology Platform pathologists), the modal (most frequently observed values) scores were added, resulting in a set of reference images (S1). These reference images were available for the second round reading of the still blinded external quality assessment tissue microarray slides.
PD-L1 immunohistochemical scoring in the Lungscape cohort
PD-L1 immunohistochemistry staining of European Thoracic Oncology Platform tumor tissue microarray sections was performed in three laboratories according to DAKO PD-L1 immunohistochemistry 28-8 PharmDx™ manufacturer’s protocol. PD-L1 stained tissue microarray slides were returned to the original centers for reading.
PD-L1 immunohistochemistry scores were recorded as both, percent of tumor cell membrane staining for each core separately, and an overall score for all cores combined. Tissue microarray sections, with up to four cores per case, were scored. A six-level categorization was considered for PD-L1 immunohistochemical tissue microarray results: “<1%”, “1–5%”, “5–10%”, “10–25%”, “25–50%”, “≥50%”, while three different PD-L1 overexpression cutoffs were used: 1, 50, and 25%, with the first two being of primary interest.
Cross-validation of PD-L1 analysis between tissue microarrays and whole sections
Scoring of PD-L1 on whole sections was performed in 10% randomly selected samples of all Lungscape cases. Samples were derived proportionally from ten participating centers, using a stratified sampling algorithm within each center. Histology (adenocarcinoma, other) and tissue microarray PD-L1 status were the stratification factors considered. The same six-level categorization was applied for PD-L1 expression determination by whole sections, as with tissue microarrays. Reading of each whole section and corresponding tissue microarray core was performed by the same local pathologist.
Statistical analysis
External quality assessment
Using as benchmark values the most frequent values observed (mode values), the agreement between the PD-L1 scoring assigned by the centers’ pathologists and the scoring were calculated and agreement rates for each external quality assessment round are presented. A five-level grouping was considered for scoring PD-L1 (<1%”, “1 -< 5%”, “5 -< 10%”, “10 -<50%”, “≥50%”, Figs. 1–2).
Tissue microarray core variability
Variability between the (≤4) tissue microarray cores of each case, is assessed by core agreement on the clinically relevant two-level categorization, according to the specific cutoffs (primarily 1% and 50% and secondarily 25%). Complete core agreement is achieved when available cores are all either below the selected cutoff or not (not evaluable cases excluded).
Cross-validation of tissue microarray and whole section
Sensitivity and specificity of the tissue microarray method (using the corresponding ‘whole section’ result as gold standard) is estimated overall and separately for the two primary histologies (adenocarcinoma/squamous cell), as well as by the number of cores available and by the core agreement status (complete/noncomplete). In addition, Cohen’s Kappa coefficient along with 95% confidence interval (CI) is used to measure the degree of agreement between tissue microarray and whole section methods (not evaluable cases treated as missing).
Results
Analysis cohorts
A total of 2402 retrospective cases from 15 centers have been captured in the European Thoracic Oncology Platform Lungscape database, of which 2182 have PD-L1 immunohistochemical information. Evaluable immunohistochemical tissue microarray overall result is recorded for 2008 cases (cohort-1), while 174 samples are deemed as “Not Evaluable” and are excluded from the analysis. PD-L1 information on whole sections is available for 237 cases (cohort-2).
Prestudy external quality assessment tissue microarray reading
Three months after the initial training, an independent, blinded, tissue microarray of 12 cases (first round) revealed an agreement rate of 73.3% between pathologists (PD-L1 score distributed in five levels, Fig. 1). As some of the cores were scored with more than one level, a set of reference images was made (S1). The same external quality assessment tissue microarray slide was read again 3 months later (second round). Reference images were available, and the agreement rate reached 80.8% (Fig. 2).
European Thoracic Oncology Platform study tissue microarray core variability
In cohort-1, almost half of the cases (49.6%) had available information on all four cores. When considering the 1% cutoff, the concordant cases represent 83.0% of cohort-1, respective percentage for the 25% cutoff is 89.1%. Highest percentage of concordant cases is observed for the 50% cutoff: 91.5% (Table 1). The highest percentage of cases in agreement, using the 1% cutoff is observed for the subgroup with available information on two cores (90.6%), followed by the three (85.2%) and then four cores (79.6%). A similar pattern but with higher agreement rates is observed for 25% (two cores: 94.8%, three cores: 88.7%, four cores: 87.4%) and 50% cutoffs (two cores: 94.8%, three: 90.7%, four: 90.8%).
A measure of intratumoral heterogeneity is obtained by comparing the minimum vs maximum PD-L1 immunohistochemical tumor cell expression of the cores available for each tumor case (Table 2). Highest and lowest immunohistochemical PD-L1 core scores perfectly match for 1198 (69.5%) of the 1724 cases. The remaining 30.5% are discrepant cases, with the largest difference between the minimum and maximum core result in PD-L1 expression (<1% vs ≥50%) observed in 37 out of 1724 cases (2.1%). If the core with the lowest PD-L1 expression is <1%, the highest core has 1/4 chance of expression above the 1% threshold (24.6%, 293/1188).
Cross-validation of PD-L1 tissue microarray and whole sections scoring methods
Considering the whole section score result as ‘gold standard’ for defining the PD-L1 expression, the sensitivity of the tissue microarray method for the 1, 25, and 50% cutoffs is: 80.4% (86/107), 77.8% (42/54), and 79.1% (34/43). Corresponding results for tissue microarray specificity (1%/25%/50% cutoffs) are: 89.9% (89/99), 95.4% (145/152), and 97.5% (159/163). Kappa coefficient values indicate substantial to strong agreement between the two methods for all three cutoffs. Corresponding values and 95% CIs for 1%/25%/50% cutoffs are: 0.70 (0.60–0.80), 0.75 (0.65–0.86) and 0.80 (0.70–0.90) (Figs. 3–5).
Respective results by number of cores available are presented in Table S1. For the 1% cutoff higher sensitivity is observed when three and four cores are available: 81.3% (13/16) and 79.2% (42/53). For 50% highest sensitivity and specificity are detected when four cores are available: 87.0% (20/23), 100.0% (85/85).
Results for sensitivity, specificity of the tissue microarray method and Kappa coefficient by histology are presented in Figs. S1–6.
Agreement of PD-L1 score result based upon tissue microarrays and whole sections is illustrated in Table 3. Complete agreement between the two methods is achieved for 60.0% (142/237) of the cases, while 17.7% of the cases (42/237) are underestimated, 9.3% (22/237) overestimated, and 12.2% (29/237) were not evaluable by tissue microarrays, but evaluable by whole sections (six level PD-L1 categorization).
Sensitivity of the tissue microarray method is lower when complete core agreement is achieved for the 1% cutoff compared to noncomplete (Table S2). The opposite holds for 25 and 50%.
Discussion
This study represents the largest PD-L1 stained cohort, confirming PD-L1 heterogeneity in non-small cell lung carcinoma. The effect of sampling, as measured by the sensitivity of tissue microarray cores compared with whole sections, is, for the relevant thresholds, in the 80% range. In addition, the results support the need for training and external quality assessment in predictive testing as well as demonstrating the value of reference images to guide threshold settings during training and alignment.
Conceptually the issue of PD-L1 intratumor heterogeneity may be approached in two ways. Firstly, by analysis of the resection specimen and samples thereof (either tissue microarray or fields of view), which are assumed to be representative for biopsies: tissue microarray cores are similar in size to transbronchial biopsy samples. The second option for examining the heterogeneity of PD-L1 expression is by comparing different samples obtained in vivo of the same tumor. Both approaches are summarized in Table S3. Other studies compared <30 patients [10, 11] or had insufficient detailed data [12, 13]. The range of reported concordance rates, for the 1 and 50% thresholds, is more or less similar (79–97%). Our data fit in the lower range, but derived from considerably more cases. The reported methods of investigation for the tissue microarray approach varied greatly, and most studies did not report over- or underestimation. In our study there is 17.7% underestimation and 9.3% overestimation of the PD-L1 expression by tissue microarray scores. This is in line with another study [14] and a recent model study, where a fraction of 10% underscoring at the 1% threshold was calculated for samples of small biopsy size [15].
Two other studies reported (Table S3) PD-L1 tumor positivity scores for two sampling approaches [16, 17]. Interestingly, in these smaller studies (<100 cases) cases with samples with a PD-L1 score >50% could also have a sample where a lower (<50%) PD-L1 score was possible. Notable, however, was the fact that where there was a sample score of <1%, another sample score for the same tumor never exceeded 50%. However, in our study, with a >20 times larger sample size, the variance in PD-L1 expression including <1% and >50% samples within a case was present in 2.1% of all cases. This wide and clinically significant variation in PD-L1 scoring cores from the same tumor is therefore likely to be uncommon in clinical practice, but the small risk should be acknowledged. Three different studies report on underestimation of PD-L1 status in small samples, perhaps therefore explaining the finding in previous clinical phase-III studies for the 9–10% responders in PD-L1 (possibly false) negative non-small cell lung carcinoma patients [18,19,20]. In our study, we also note that of all the cases scored ≥50% by whole section, 18% were underscored on the tissue microarray, although 44% of these underscored cases were in the adjacent (≥25–49%) category. Similarly, of the cases scored <1% by whole section, 9% were ‘over-scored’ but half of these were in the adjacent 1–5% group. This may have implications for how pathologists approach cases where the score is just above or below a clinically significant category.
Issue number of biopsies
As heterogeneity of PD-L1 expression is present in non-small cell lung carcinoma, the question arises as to what the optimal samples size (number) is [5]. Few studies addressed this issue. One sample is not enough: 1 negative core (PD-L1 < 1%) still had a 26% chance on a positive score, if up to four cores were taken [21]. All the immunohistochemistry protocols require a minimum of 100 viable tumor cells for PD-L1 [17]. In this context, Sakakibara et al. demonstrated, for comparison that in cytology samples 100–1000 vs >2000 tumor cells showed an increase in correlation coefficient from 0.19 (p = 0.49; n = 15) to 0.68 (p = 0.0019; n = 18) when more than 2000 tumor cells were examined [12]. In a retrospective study by Ilie et al., where the number of biopsy cores varied between 1 and 12 (mean 4), a trend toward a significant difference was present between the average number of biopsy fragments in discordant vs concordant cases (3.4 vs 6.8; p = 0.07) [14]. Munari et al. found for the 50% PD-L1 threshold an optimum at three cores (out of a maximum of 5) [22]. For the 1% threshold the optimum was less clear with the optimum at five out of five cores. In our study for the 1% cutoff the highest sensitivity is observed when either three or four cores are available. For the 50% cutoff highest sensitivity is observed when four (out of four) cores are available, suggesting that at least four biopsies need to be taken. These studies use, for the comparison of small samples, a single tissue block of a resection specimen. This is probably reasonable for a ‘gold standard’ as the spatial distribution of heterogeneity of PD-L1 expression, examined by comparing multiple blocks, seems to be represented by a single block in most cases [23, 24]. In the discussion, core size and biopsy size need to be taken into account, something which has not been addressed so far. A 19 gauge (inner diameter 0.69 mm) transthoracic needle biopsy of 1 cm is ~6–8 times larger than a bronchial biopsy. Overall, the optimum number of biopsy type of samples is not determined, but four seems to be the minimum. Besides sample size, the number of tumor cells per sample plays a role. In bronchial biopsies the range of tumor cells varies from 94%, down to as little as to 0.1% in some cases [25, 26]. In this respect, the minimum number of 100 tumor cells, that needs to be present for a valid PD-L1 score, is probably valid in homogenous samples, but heterogeneity is likely to be better covered with more tumor cells (>2000) [12]. In search for a balance between practical tissue sampling and an optimal PD-L1 scoring approach the minimum number of 100 tumor cells is a compromise, that will not be adequate in all cases with heterogeneity for PD-L1 expression: an issue that cannot be known based of small biopsy samples alone.
In future clinical trials, recording of biopsy size and tumor load may permit further refinement with increase of predictive accuracy. In practical terms, however, it is often difficult to control or determine the amount of tumor in clinical samples. It is clear that a larger tumor sample is more likely to give a more accurate score for the patient’s disease burden. In practice, these data emphasize the importance of obtaining as much tumour as possible, within the constraints of patient safety, and the possibility of inaccurate PD-L1 scoring when small amounts of tumour are scored. The 100 cell minimum is likely to be below the optimum for accurate scoring. Heterogeneity is likely to be a significant contributor to inaccurate PD-L1 scoring and a plausible explanation for ‘unexpected’ clinical responses in PD-L1 negative cases. Moreover, sampling more and larger biopsies is likely to avoid to a large extend the false negative rate of PD-L1 testing.
Limitations of the study
As the study is performed on resection specimen the PD-L1 positivity rate is a reflection of early stage disease, rather than extended disease. Due to the limited sample size in cases with one core and three cores available, inference should be treated with caution. A possible effect of preanalytical variables is not excluded [27]. Moreover, the use of cores instead of biopsy samples is an approximation of clinical practice. However, tissue microarray cores permit the examination of more non-small cell lung carcinoma cases in a less invasive manner.
Conclusion
In conclusion, we demonstrate that classification of PD-L1 on small biopsy samples does not represent the overall expression of PD-L1 in all non-small cell lung carcinoma cases, although the majority of cases are ‘correctly’ classified. Underestimation of PD-L1 expression on small samples is more common than overestimation and the former may be the explanation for clinical responses observed in clinical trials based on apparently PD-L1 negative biopsies. In future clinical trials, sampling more and larger biopsies, recording of biopsy size and tumor load may permit further refinement with increase of predictive accuracy.
References
Melosky B, Chu Q, Juergens R, Leighl N, McLeod D, Hirsh V. Pointed progress in second-line advanced non-small-cell lung cancer: the rapidly evolving field of checkpoint inhibition. J Clin Oncol. 2016;34:1676–88.
Assi HI, Kamphorst AO, Moukalled NM, Ramalingam SS. Immune checkpoint inhibitors in advanced non-small cell lung cancer. Cancer. 2018;124:248–61.
Gong J, Chehrazi-Raffle A, Reddi S, Salgia R. Development of PD-1 and PD-L1 inhibitors as a form of cancer immunotherapy: a comprehensive review of registration trials and future considerations. J Immunother Cancer. 2018;6:8.
Büttner R, Gosney JR, Skov BG, Adam J, Motoi N, Bloom KJ, et al. Programmed death-ligand 1 immunohistochemistry testing: a review of analytical assays and clinical implementation in non-small-cell lung cancer. J Clin Oncol. 2017;35:3867–76.
Nakamura S, Hayashi K, Imaoka Y, Kitamura Y, Akazawa Y, Tabata K, et al. Intratumoral heterogeneity of programmed cell death ligand-1 expression is common in lung cancer. PLoS ONE. 2017;12:e0186192.
Peters S, Weder W, Dafni U, Kerr KM, Bubendorf L, Meldgaard P, et al. Lungscape: resected non-small-cell lung cancer outcome by clinical and pathological parameters. J Thorac Oncol. 2014;9:1675–84.
Kerr KM, Dafni U, Schulze K, Thunnissen E, Bubendorf L, Hager H, et al. Prevalence and clinical association of gene mutations through multiplex mutation testing in patients with NSCLC: Results from the ETOP Lungscape Project. Ann Oncol 2018;29. https://doi.org/10.1093/annonc/mdx629.
Blackhall FH, Peters S, Bubendorf L, Dafni U, Kerr KM, Hager H, et al. Prevalence and clinical outcomes for patients with ALK-positive resected stage I to III adenocarcinoma: results from the European Thoracic Oncology Platform Lungscape Project. J Clin Oncol. 2014;32:2780–87.
Kerr KM, Thunnissen E, Dafni U, Finn SP, Bubendorf L, Soltermann A, et al. A retrospective cohort study of PD-L1 prevalence, molecular associations and clinical outcomes in patients with NSCLC: results from the European Thoracic Oncology Platform (ETOP) Lungscape Project. Lung Cancer 2019;131:95–103.
Heymann JJ, Bulman WA, Swinarski D, Pagan CA, Crapanzano JP, Haghighi M, et al. PD-L1 expression in non-small cell lung carcinoma: comparison among cytology, small biopsy, and surgical resection specimens. Cancer Cytopathol. 2017;125:896–907.
Stoy SP, Rosen L, Mueller J, Murgu S. Programmed death-ligand 1 testing of lung cancer cytology specimens obtained with bronchoscopy. Cancer Cytopathol. 2018;126:122–8.
Sakakibara R, Inamura K, Tambo Y, Ninomiya H, Kitazono S, Yanagitani N, et al. EBUS-TBNA as a promising method for the evaluation of tumor PD-L1 expression in lung cancer. Clin Lung Cancer. 2017;18:527–34:e1.
McLaughlin J, Han G, Schalper KA, Carvajal-Hausdorf D, Pelekanou V, Rehman J, et al. Quantitative assessment of the heterogeneity of PD-L1 expression in non-small-cell lung cancer. JAMA Oncol. 2016;2:46–54.
Ilie M, Long-Mira E, Bence C, Butori C, Lassalle S, Bouhlel L, et al. Comparative study of the PD-L1 status between surgically resected specimens and matched biopsies of NSCLC patients reveal major discordances: a potential issue for anti-PD-L1 therapeutic strategies. Ann Oncol. 2016;27:147–53.
Bigras G, Mairs S, Swanson PE, Morel D, Lai R, Izevbaye I. Small biopsies misclassify up to 35% of PD-L1 assessments in advanced lung non–small cell lung carcinomas. Appl Immunohistochem Mol Morphol 2018;10:701–8.
Gniadek TJ, Li QK, Tully E, Chatterjee S, Nimmagadda S, Gabrielson E. Heterogeneous expression of PD-L1 in pulmonary squamous cell carcinoma and adenocarcinoma: implications for assessment by small biopsy. Mod Pathol. 2017;30:530–8.
Skov BG, Skov T. Paired comparison of PD-L1 expression on cytologic and histologic specimens from malignancies in the lung assessed with PD-L1 IHC 28-8pharmDx and PD-L1 IHC 22C3pharmDx. Appl Immunohistochem Mol Morphol. 2017;25:453–9.
Gandhi L, Rodríguez-Abreu D, Gadgeel S, Esteban E, Felip E, De Angelis F, et al. Pembrolizumab plus chemotherapy in metastatic non–small-cell lung cancer. N Engl J Med. 2018;378:2078–92.
Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372:2018–28.
Horn L, Spigel DR, Vokes EE, Holgado E, Ready N, Steins M, et al. Nivolumab versus docetaxel in previously treated patients with advanced non-small-cell lung cancer: two-year outcomes from two randomized, open-label, phase III trials (CheckMate 017 and CheckMate 057). J Clin Oncol. 2017;35:3924–33.
Gagné A, Enlow W, Pigeon MA, Orain M, Turcotte S, Bossé Y, et al. Comprehensive assessment of PD-L1 staining heterogeneity in pulmonary adenocarcinomas using tissue microarrays. Am J Surg Pathol. 2018;42:687–94.
Munari E, Zamboni G, Lunardi G, Marchionni L, Marconi M, Sommaggio M, et al. PD-L1 expression heterogeneity in non-small cell lung cancer: defining criteria for harmonization between biopsy specimens and whole sections. J Thorac Oncol 2018;13:1113–20.
Rehman JA, Han G, Carvajal-Hausdorf DE, Wasserman BE, Pelekanou V, Mani NL, et al. Quantitative and pathologist-read comparison of the heterogeneity of programmed death-ligand 1 (PD-L1) expression in non-small cell lung cancer. Mod Pathol. 2017;30:340–9.
Scorer P, Scott M, Lawson N, Ratcliffe MJ, Barker C, Rebelatto MC, et al. Consistency of tumor and immune cell programmed cell death ligand-1 expression within and between tumor blocks using the VENTANA SP263 assay. Diagn Pathol. 2018;13:1–10.
Coghlin CL, Smith LJ, Bakar S, Stewart KN, Devereux GS, Nicolson MC, et al. Quantitative analysis of tumor in bronchial biopsy specimens. J Thorac Oncol. 2010;5:448–52.
Thunnissen E, Kerr KM, Herth FJ, Lantuejoul S, Papotti M, Rintoul RC, et al. The challenge of NSCLC diagnosis and predictive analysis on small samples. Practical approach of a working group. Lung Cancer. 2012;76:1–18.
van Seijen M, Brcic L, Gonzales AN, Sansano I, Bendek M, Brcic I, et al. ETOP Impact of delayed and prolonged fixation on the evaluation ofimmunohistochemical staining on lung carcinoma resection specimen. Virchows Arch 2019;475:191–9.
European Thoracic Oncology Platform Lungscape Consortium
Rolf A. Stahel21, Rafael Rosell22, Fiona Blackhall23,24, Urania Dafni3,25, Keith M. Kerr2,26, Miguel Angel Molina27, Lukas Bubendorf4, Walter Weder28,29, Erik Thunnissen1,30, Solange Peters20, Stephen Finn5,31, Anita Hiltbrunner32, Roswitha Kammler32, Thomas Geiger32, Nesa Marti32, Zoi Tsourti25, Varvara Polydoropoulou25, Panagiota Zygoura25, Marianne Nicolson26, David A. J. Stevenson26, William Mathieson26, Egbert Smit30, Teodora Radonic30, Alex Soltermann29, Undine Rulle29, Alessandra Curioni29, Steven G. Gray31, Kathy Gately31, Martin Barr31, Peter Meldgaard33, Line B. Madsen33, Spasenija Savic34, Didier Lardinois34, Kristiaan Nackaerts35, Christophe Dooms35, Els Wauters35, Sara Van Der Borght35, Wojciech Biernat36, Ania Wrona36, Witold Rzyman36, Jacek Jassem36, Hendrik Dienemann37, Thomas Muley37, Arne Warth37, Antonio Marchetti38, Graziano De Luca38, Alessia di Lorito38, Anne-Marie Dingemans39, Ernst-Jan M. Speel39, Andrea Ruland39, Saraswati Pokharel40, Richard Cheney40, Philip Ferenczy40, Anne Marie Quinn24, Lynsey Franklin24, Paul Baas41, Kim Monkhorst41, Bart van de Wiel41, Carlos Camps42, Miguel Martorell42, Atilio Navarro42
Funding
The European Thoracic Oncology Platform received financial support for study conduct from Bristol-Myers Squibb. The Lungscape 005 PD-L1 study, including the 2-day DAKO training, was funded by Bristol-Meyers Squibb.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Conflict of interest
LB: advisory boards from Bristol-Myers Squibb. Other authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Thunnissen, E., Kerr, K.M., Dafni, U. et al. Programmed death-ligand 1 expression influenced by tissue sample size. Scoring based on tissue microarrays’ and cross-validation with resections, in patients with, stage I–III, non-small cell lung carcinoma of the European Thoracic Oncology Platform Lungscape cohort. Mod Pathol 33, 792–801 (2020). https://doi.org/10.1038/s41379-019-0383-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-019-0383-9
This article is cited by
-
Nationwide differences in cytology fixation and processing methods and their impact on interlaboratory variation in PD-L1 positivity
Virchows Archiv (2023)
-
Diagnosis of atypical carcinoid can be made on biopsies > 4 mm2 and is accurate
Virchows Archiv (2022)
-
Predictive biomarkers for response to immune checkpoint inhibitors in lung cancer: PD-L1 and beyond
Virchows Archiv (2021)
-
PD-L1 amplification is associated with an immune cell rich phenotype in squamous cell cancer of the lung
Cancer Immunology, Immunotherapy (2021)
-
Detection of programmed cell death-ligand 1 using 22C3 antibody in patients with unresectable stage III non-small cell lung cancer receiving chemoradiotherapy
International Journal of Clinical Oncology (2021)