Abstract
Key message
We developed a machine learning-based model to identify the hidden labels of m6A candidates from noisy m6A-seq data.
Abstract
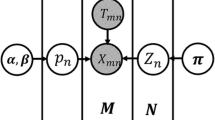
Peak-calling approaches, such as MeRIP-seq or m6A-seq, are commonly used to map m6A modifications. However, these technologies can only map m6A sites with 100–200 nt resolution and cannot reveal the precise location or the number of modified residues in a transcript. To address this challenge, we developed a novel machine learning-based approach, named HLMethy, to assign labels to m6A candidates from noisy m6A-seq data. The multiple instance learning framework was adopted and two different training strategies were used to generate the classification model. To test the performance of our model, the m6A sites with single-base resolution were used and our model achieved comparable performance against existing instance-level predictors, which suggest that our model has the potential to improve the data quality of m6A-seq at reduced costs. What’s more, our generic framework can be extended to other newly found modifications that are found by peak-calling approaches. The source code of HLMethy is available at https://github.com/liuze-nwafu/HLMethy.





Similar content being viewed by others
References
Andrews S, Hofmann T, Tsochantaridis I (2002) Multiple instance learning with generalized support vector machines. In: Eighteenth national conference on Artificial intelligence
Babenko B (2008) Multiple instance learning: algorithms and applications
Chang CC, Lin CJ (2011) LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol 27:1–27. https://doi.org/10.1145/1961189.1961199
Chen W, Feng PM, Ding H et al (2015a) iRNA-methyl: identifying N6-methyladenosine sites using pseudo nucleotide composition. Anal Biochem 490(1):26–33. https://doi.org/10.1016/j.ab.2015.08.021
Chen W, Hong T, Liang ZY et al (2015b) Identification and analysis of the N6-methyladenosine in the Saccharomyces cerevisiae transcriptome. Sci Rep 5:13859. https://doi.org/10.1038/srep13859
Chen W, Tang H, Lin H (2016a) MethyRNA: a web server for identification of N6-methyladenosine sites. J Biomol Struct Dyn 35(3):683–687. https://doi.org/10.1080/07391102.2016.1157761
Chen W, Feng PM, Ding H, Lin H et al (2016b) Identifying N (6)-methyladenosine sites in the Arabidopsis thaliana transcriptome. Mol Genet Genomics 291:2225–2229. https://doi.org/10.1007/s00438-016-1243-7
Chen W, Yang H, Feng P, Ding H, Lin H (2017) iDNA4mC: identifying DNA N4-methylcytosine sites based on nucleotide chemical properties. Bioinformatics 33(22):3518–3523. https://doi.org/10.1093/bioinformatics/btx479
Chen KQ, Wei Z, Zhang Q et al (2019) WHISTLE: a high-accuracy map of the human N6-methyladenosine (m6A) epitranscriptome predicted using a machine learning approach. Nucleic Acid Res 47(7):e41. https://doi.org/10.1093/nar/gkz074
Delatte B, Wang F, Ngoc LV et al (2016) Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science 351(6270):282–285. https://doi.org/10.1126/science.aac5253
Desrosiers R, Friderici K, Rottman F (1974) Identification of methylated nucleosides in messenger rna from novikoff hepatoma cells. Proc Natl Acad Sci USA 71(10):3971–3975. https://doi.org/10.1073/pnas.71.10.3971
Dominissini D, Sharon MM, Schwartz S et al (2012) Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485(7397):201–206. https://doi.org/10.1038/nature11112
Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S et al (2016) The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature 530(7591):441–446. https://doi.org/10.1038/nature16998
Edelheit S, Schwartz S, Mumbach MR, Wurtzel O, Sorek R (2013) Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet 9(6):e1003602. https://doi.org/10.1371/journal.pgen.1003602
Eksi R, Li HD, Menon R et al (2013) Systematically differentiating functions for alternatively spliced isoforms through integrating RNA-seq data. PLoS Comput Biol 9(11):e1003314. https://doi.org/10.1371/journal.pcbi.1003314
Feng PM, Yang H, Ding H, Lin H, Chen W, Chou KC (2018) iDNA6 mA-PseKNC: identifying DNA N6-methyladenosine sites by incorporating nucleotide physicochemical properties into PseKNC. Genomics 111(1):96–102. https://doi.org/10.1016/j.ygeno.2018.01.005
Göke J, Schulz MH, Lasserre J, Vingron M (2012) Estimation of pairwise sequence similarity of mammalian enhancers with word neighbourhood counts. Bioinformatics 28(5):656–663. https://doi.org/10.1093/bioinformatics/bts028
Grozhik AV, Jaffrey SR (2018) Distinguishing RNA modifications from noise in epitranscriptome maps. Nat Chem Biol 14(3):215–225. https://doi.org/10.1038/nchembio.2546
Huang Y, Niu B, Gao Y, Fu LM, Li WZ (2010) CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 26(5):680–682. https://doi.org/10.1093/bioinformatics/btq003
Legrand C, Tuorto F, Hartmann M et al (2017) Statistically robust methylation calling for whole-transcriptome bisulfite sequencing reveals distinct methylation patterns for mouse RNAs. Genome Res 27:1589–1596. https://doi.org/10.1101/gr.210666.116
Li Z, Chen YX, Mu DS et al (2012) Comparison of the two major classes of assembly algorithms: overlap-layout-consensus and de-bruijn-graph. Brief Funct Genomics 11(1):25–37. https://doi.org/10.1093/bfgp/elr035
Li X, Xiong X, Wang K, Wang L, Yi C (2016a) Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat Chem Biol 12(5):311. https://doi.org/10.1038/nchembio.2040
Li GQ, Liu Z, Shen HB et al (2016b) Target M6A: identifying N6-methyladenosine sites from RNA sequences via position-specific nucleotide propensities and a support vector machine. IEEE Trans Nanobiosci 15(7):674–682. https://doi.org/10.1109/TNB.2016.2599115
Linder B, Grozhik AV, Anthony O et al (2015) Single-nucleotide resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods 12(8):767–772. https://doi.org/10.1038/nmeth.3453
Liu Z, Xiao X, Yu DJ et al (2016) pRNAm-PC: predicting N6-methyladenosine sites in RNA sequences via physical–chemical properties. Anal Biochem 497:60–67. https://doi.org/10.1016/j.ab.2015.12.017
Luo GZ, Alice MQ, Zheng GQ et al (2014) Unique features of the m6A methylome in Arabidopsis thaliana. Nat Commun 5:5630. https://doi.org/10.1038/ncomms6630
Meyer KD, Jaffrey SR (2014) The dynamic epitranscriptome: n6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol 15(5):313–326. https://doi.org/10.1038/nrm3785
Minhas F, Ben-Hur A (2012) Multiple instance learning of calmodulin binding sites. Bioinformatics 28(18):i416–i422. https://doi.org/10.1093/bioinformatics/bts416
Minhas F, Ross ED, Ben-Hur A (2017) Amino acid composition predicts prion activity. Comput Biol 13(4):e1005465. https://doi.org/10.1371/journal.pcbi.1005465
Pelckmans K, Suykens JAK, Moor BD (2006) Additive regularization trade-off: fusion of training and validation levels in Kernel methods. Mach Learn 62(3):217–252. https://doi.org/10.1007/s10994-005-5315-x
Safra M, Sas-chen A, Nir R et al (2017) The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 551:251–255. https://doi.org/10.1038/nature24456
Schwartz SD, Agarwala MR, Mumbach M et al (2013) High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 155(6):1409–1421. https://doi.org/10.1016/j.cell.2013.10.047
Schwartz S, Bernstein D, Mumbach M et al (2014) Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159(1):148–162. https://doi.org/10.1016/j.cell.2014.08.028
Shen L, Liang Z, Gu XF et al (2016) N(6)-methyladenosine RNA modification regulates shoot stem cell fate in arabidopsis. Dev Cell 38(2):186–200. https://doi.org/10.1016/j.devcel.2016.06.008
Song J, Zhai JJ, Bian E, Song YJ, Yu JT, Ma C (2018) Transcriptome-wide annotation of m5C RNA modifications using machine learning. Front Plant Sci 9:519. https://doi.org/10.3389/fpls.2018.00519
Squires JE, Patel HR, Marco N et al (2012) Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res 40(11):5023–5033. https://doi.org/10.1093/nar/gks144
Wang X, Yan R (2018) RFAthM6A: a new tool for predicting m(6)A sites in Arabidopsis thaliana. Plant Mol Biol 96(3):327–337. https://doi.org/10.1007/s11103-018-0698-9
Xiang SN, Liu K, Yan ZM et al (2016a) RNAMethPre: a web server for the prediction and query of mRNA m6A sites. PLoS ONE 11(10):e0162707. https://doi.org/10.1371/journal.pone.0162707
Xiang SN, Yan ZM, Liu K, Zhang Y, Sun Z (2016b) AthMethPre: a web server for the prediction and query of mRNA m(6)A sites in Arabidopsis thaliana. Mol BioSyst 12(11):3333–3337. https://doi.org/10.1039/C6MB00536E
Xing PW, Su R, Guo F, Wei LY (2017) Identifying N(6)-methyladenosine sites using multi-interval nucleotide pair position specificity and support vector machine. Sci Rep 7:46757. https://doi.org/10.1038/srep46757
Xu Y, Li Y, Shen Z et al (2017) Parallel multiple instance learning for extremely large histopathology image analysis. BMC Bioinform 18(1):360. https://doi.org/10.1186/s12859-017-1768-8
Zhai JJ, Song J, Cheng Q, Tang Y, Ma C (2018) PEA: an integrated R toolkit for plant epitranscriptome analysis. Bioinformatics 34(21):3747–3749. https://doi.org/10.1093/bioinformatics/bty421
Zhang CX, Chen YS, Sun BF et al (2017) m(6)A modulates haematopoietic stem and progenitor cell specification. Nature 549:273–276. https://doi.org/10.1038/nature23883
Zhang SY, Zhang SW, Fan XN et al (2019) Global analysis of N6-methyladenosine functions and its disease association using deep learning and network-based methods. PLoS Comput Biol 15(1):e1006663. https://doi.org/10.1371/journal.pcbi.1006663
Zhou Y, Zeng P, Li YH et al (2016) SRAMP: prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res 44(10):e91. https://doi.org/10.1093/nar/gkw104
Acknowledgements
This work was supported by the Start-up fund of Northwest A&F University (Z109021809), National Natural Science Foundation of China (61902323), and Postdoctoral Research Foundation of China (2018M643744).
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: ZL, WD. Performed the experiments: ZL, WJ. Analyzed the data: QWL, ZLH. Contributed reagents/materials/analysis tools: WD, WJ. Contributed to the writing of the manuscript: ZL, WJL, WJ, ZLH. Designed and developed the software: ZL, WJL, QWL.
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Liu, Z., Dong, W., Luo, W. et al. HLMethy: a machine learning-based model to identify the hidden labels of m6A candidates. Plant Mol Biol 101, 575–584 (2019). https://doi.org/10.1007/s11103-019-00930-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-019-00930-x