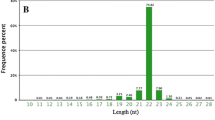
Abstract
Heart development is a complex process regulated by multi-layered genetic as well epigenetic regulators many of which are still unknown. Besides their critical role during cardiac development, these molecular regulators emerge as key modulators of cardiovascular pathologies, where fetal cardiac genes’ re-expression is witnessed. MicroRNAs have recently emerged as a crucial part of signalling cascade in both development and diseases. We aimed to identify, validate, and perform functional annotation of putative novel miRNAs using chicken as a cardiac development model system. Novel miRNAs were obtained through deep sequencing of small RNAs extracted from chicken embryonic cardiac tissue of different developmental stages. After filtering out real pre-miRNAs, their expression analysis, potential target gene’s prediction and functional annotations were performed. Expression analysis revealed that miRNAs were differentially expressed during different developmental stages of chicken heart. The expression of selected putative novel miRNAs was further validated by real-time PCR. Our analysis indicated the presence of novel cardiac miRNAs that might be regulating critical cardiac development events such as cardiac cell growth, differentiation, cardiac action potential generation and signal transduction.




Similar content being viewed by others
References
Srivastava D, Olson EN (2000) A genetic blueprint for cardiac development. Nature 407:221–226
Abu-Issa R, Kirby ML (2007) Heart field: from mesoderm to heart tube. Annu Rev Cell Dev Biol 23:45–68
Nandi SS, Mishra PK (2015) Harnessing fetal and adult genetic reprograming for therapy of heart disease. J Nat Sci 1:4–6
Barry SP, Davidson SM, Townsend PA (2008) Molecular regulation of cardiac hypertrophy. Int J Biochem Cell Biol 40:2023–2039
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Fuller A, Qian L (2014) MiRiad roles for MicroRNAs in cardiac development and regeneration. Cells 3:724–750
Rao PK, Toyama Y, Chiang HR et al (2009) Loss of cardiac microRNA-mediated regulation leads to dilated cardiomyopathy and heart failure. Circ Res 105:585–594
Rustagi Y, Jaiswal HK, Rawal K et al (2015) Comparative characterization of cardiac development specific microRNAs: fetal regulators for future. PLoS ONE 10:e0139359
Epstein JA (2010) Cardiac development and implications for heart disease. N Engl J Med 363:1638–1647
Katz MG, Fargnoli AS, Kendle AP et al (2016) The role of microRNAs in cardiac development and regenerative capacity. Am J Physiol 310:H528–H541
Wittig J, Münsterberg A (2016) The early stages of heart development: insights from chicken embryos. J Cardiovasc Dev Dis 3:12
Gao D, Middleton R, Rasko JEJ et al (2013) MiREval 2.0: a web tool for simple microRNA prediction in genome sequences. Bioinformatics 29:3225–3226
Jiang P, Wu H, Wang W et al (2007) MiPred: classification of real and pseudo microRNA precursors using random forest prediction model with combined features. Nucleic Acids Res 35:W339–W344
Gruber AR, Lorenz R, Bernhart SH et al (2008) The Vienna RNA websuite. Nucleic Acids Res 36:W70–W74
Kozomara A, Griffiths-Jones S (2014) MiRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42:D68–D73
Wong N, Wang X (2015) miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 43:D146–D152
Mi H, Muruganujan A, Thomas PD (2013) PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res 41:D377–D386
Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57
Shannon P, Markiel A, Ozier O et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504
Varkonyi-Gasic E, Wu R, Wood M et al (2007) Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3:12
Yang LH, Wang SL, Tang LL et al (2014) Universal stem-loop primer method for screening and quantification of microRNA. PLoS ONE 9:e115293
Laganà A, Veneziano D, Spata T et al (2015) Identification of general and heart-specific miRNAs in sheep (Ovis aries). PLoS ONE 10:e0143313
Ambros V, Bartel B, Bartel DP et al (2003) A uniform system for microRNA annotation. RNA 9:277–279
Martinsen BJ (2005) Reference guide to the stages of chick heart embryology. Dev Dyn 233:1217–1237
Xu H, Wang X, Du Z, Li N (2006) Identification of microRNAs from different tissues of chicken embryo and adult chicken. FEBS Lett 580:3610–3616
Ji Z, Wang G, Xie Z et al (2012) Identification and characterization of microRNA in the dairy goat (Capra hircus) mammary gland by Solexa deep-sequencing technology. Mol Biol Rep 39:9361–9371
Ge X, Zhang Y, Jiang J et al (2012) Identification of microRNAs in Helicoverpa armigera and Spodoptera litura based on deep sequencing and homology analysis. Int J Biol Sci 9:1–15
Islam MT, Ferdous AS, Najnin RA et al (2015) High-throughput sequencing reveals diverse sets of conserved, nonconserved, and species-specific miRNAs in jute. Int J Genom 2015:125048
Kim GH (2013) MicroRNA regulation of cardiac conduction and arrhythmias. Transl Res 161:381–392
Yang B, Lin H, Xiao J et al (2007) The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med 13:486–491
Liu X, Zhang Y, Du W et al (2016) MiR-223-3p as a novel MicroRNA regulator of expression of voltage-gated K + channel Kv4.2 in acute myocardial infarction. Cell Physiol Biochem 39:102–114
Yang WM, Jeong HJ, Park SW, Lee W (2015) Obesity-induced miR-15b is linked causally to the development of insulin resistance through the repression of the insulin receptor in hepatocytes. Mol Nutr Food Res 59:2303–2314
Motohashi N, Alexander MS, Shimizu-Motohashi Y et al (2013) Regulation of IRS1/Akt insulin signaling by microRNA-128a during myogenesis. J Cell Sci 126:2678–2691
Min KH, Yang WM, Lee W (2018) Saturated fatty acids-induced miR-424–5p aggravates insulin resistance via targeting insulin receptor in hepatocytes. Biochem Biophys Res Commun 503:1587–1593
Ono K, Igata M, Kondo T et al (2018) Identification of microRNA that represses IRS-1 expression in liver. PLoS ONE 13:e0191553
DeBosch BJ, Muslin AJ (2008) Insulin signaling pathways and cardiac growth. J Mol Cell Cardiol 44:855–864
Jia G, Whaley-Connell A, Sowers JR (2018) Diabetic cardiomyopathy: a hyperglycaemia- and insulin-resistance-induced heart disease. Diabetologia 61:21–28
Boucher J, Kleinridders A, Ronald Kahn C (2014) Insulin receptor signaling in normal and insulin-resistant states. Cold Spring Harb Perspect Biol 6:a009191
Chen Z, Ding L, Yang W et al (2017) Hepatic activation of the FAM3C-HSF1-CaM pathway attenuates hyperglycemia of obese diabetic mice. Diabetes 66:1185–1197
Park J, Ahn S, Jayabalan AK et al (2016) Insulin signaling augments eIF4E-dependent nonsense-mediated mRNA decay in mammalian cells. Biochim Biophys Acta 1859:896–905
Tian Y, Cohen ED, Morrisey EE (2010) The importance of Wnt signaling in cardiovascular development. Pediatr Cardiol 31:342–348
Li M, Hu X, Zhu J et al (2014) Overexpression of miR-19b impairs cardiac development in zebrafish by targeting ctnnb1. Cell Physiol Biochem 33:1988–2002
Ye X, Hemida MG, Qiu Y et al (2013) MiR-126 promotes coxsackievirus replication by mediating cross-talk of ERK1/2 and Wnt/β-catenin signal pathways. Cell Mol Life Sci 70:4631–4644
Han P, Li W, Lin CH et al (2014) A long noncoding RNA protects the heart from pathological hypertrophy. Nature 514:102–106
Tian J, An X, Niu L (2017) Role of microRNAs in cardiac development and disease. Exp Ther Med 13:3–8
Ling TY, Wang XL, Chai Q et al (2017) Regulation of cardiac CACNB2 by microRNA-499: potential role in atrial fibrillation. BBA Clin 7:78–84
Rienks M, Barallobre-Barreiro J, Mayr M (2018) The emerging role of the ADAMTS family in vascular diseases. Circ Res 123:1279–1281
Liu R, Correll RN, Davis J et al (2015) Cardiac-specific deletion of protein phosphatase 1β promotes increased myofilament protein phosphorylation and contractile alterations. J Mol Cell Cardiol 87:204–213
Hoeflich A, David R, Hjortebjerg R (2018) Current IGFBP-related biomarker research in cardiovascular disease: we need more structural and functional information in clinical studies. Front Endocrinol 9:388
Acknowledgements
This study was supported by DST-SERB, Government of India (File No: EMR/2016/005914); and CSIR, Government of India (File No: 09/1132 (0004)/18-EMR-I).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11010_2019_3618_MOESM1_ESM.tif
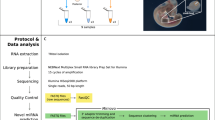
Supplementary material 1—Flowchart depicting the steps taken for identification and validation of novel miRNAs and generation of interaction network between enriched GO terms and KEEG pathways. (TIFF 148 kb)
Rights and permissions
About this article
Cite this article
Saxena, S., Mathur, P., Shukla, V. et al. Differential expression of novel MicroRNAs from developing fetal heart of Gallus gallus domesticus implies a role in cardiac development. Mol Cell Biochem 462, 157–165 (2019). https://doi.org/10.1007/s11010-019-03618-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-019-03618-4