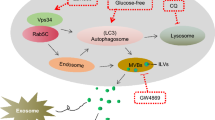
Abstract
Rasal2, a Ras-GTPase-activating protein (RasGAP), is a tumor suppressor in Luminal B breast cancer, frequently metastatic and recurrent. Exosomes (Exos) are small membrane vesicles secreted by various cell types, including tumor cells, recognized as vehicles for cell-to-cell communication. Our study aimed to investigate whether Rasal2 regulates breast cancer cell growth via affecting this process. In this paper, we described that Rasal2 knockout (KO) in MCF-7 cells enhanced exosomal release and increased autophagy-related proteins in exosomal fraction, while attenuated by exosome release inhibitor GW4869. Moreover, MCF-7 cells with chloroquine (CQ) treatment boosted Rasal2 KO-induced secretory autophagy. In addition, we presented that exosomes derived from KO MCF-7 cells (KO-exo) significantly promoted breast cancer cell proliferation compared to those from MCF-7 cells transfected with an empty crispr-cas9 plasmid serving as controls (sgNT-exo); however, exosomes purified from KO MCF-7 cells co-cultured with 3-methyladenine ((3-MA + KO)-exo)/CQ ((CQ + KO)-exo) dramatically inhibited/facilitated MCF-7 cell proliferation in contrast to KO-exo group, separately. In conclusion, our findings revealed a new mechanism of Rasal2 in the regulation of breast cancer cell proliferation via autophagy-exo-mediated pathway.
Graphic abstract





Similar content being viewed by others
References
Kamangar F, Dores GM, Anderson WF (2006) Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol 24:2137–2150
McLaughlin SK, Olsen SN, Dake B et al (2013) The RasGAP gene, RASAL2, is a tumor and metastasis suppressor. Cancer Cell 24:365–378
Olsen SN, Wronski A, Castano Z et al (2017) Loss of RasGAP tumor suppressors underlies the aggressive nature of luminal b breast cancers. Cancer Discov 7:202–217
Verweij FJ, Bebelman MP, Jimenez CR et al (2018) Quantifying exosome secretion from single cells reveals a modulatory role for GPCR signaling. J Cell Biol 217:1129–1142
Zeng Z, Li Y, Pan Y et al (2018) Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat Commun 9:5395
Li LM, Liu H, Liu XH et al (2019) Clinical significance of exosomal miRNAs and proteins in three human cancers with high mortality in China. Oncol Lett 17:11–22
Moradi-Chaleshtori M, Hashemi SM, Soudi S et al (2019) Tumor-derived exosomal microRNAs and proteins as modulators of macrophage function. J Cell Physiol 234:7970–7982
Hessvik NP, Overbye A, Brech A et al (2016) PIKfyve inhibition increases exosome release and induces secretory autophagy. Cell Mol Life Sci 73:4717–4737
Hong J, Bhat OM, Li G et al (2019) Lysosomal regulation of extracellular vesicle excretion during d-ribose-induced NLRP3 inflammasome activation in podocytes. Biochim Biophys Acta Mol Cell Res 1866:849–860
Guo H, Chitiprolu M, Roncevic L et al (2017) Atg5 disassociates the V1V0-ATPase to promote exosome production and tumor metastasis independent of canonical macroautophagy. Dev Cell 43:716–730
Miranda AM, Lasiecka ZM, Xu Y et al (2018) Neuronal lysosomal dysfunction releases exosomes harboring APP C-terminal fragments and unique lipid signatures. Nat Commun 9:291
Thery C, Zitvogel L, Amigorena S (2002) Exosomes: composition, biogenesis and function. Nat Rev Immunol 2:569–579
Jia X, Chen J, Megger DA et al (2017) Label-free proteomic analysis of exosomes derived from inducible hepatitis b Virus-Replicating HepAD38 cell line. Mol Cell Proteom 16:S144–S160
Lasoń E, Sikora E, Ogonowski J (2013) Influence of process parameters on properties of nanostructured lipid carriers (NLC) formulation. Acta Biochim Pol 60:773
Ghazizadeh E, Naseri Z, Jaafari MR et al (2018) A fires novel report of exosomal electrochemical sensor for sensing micro RNAs by using multi covalent attachment p19 with high sensitivity. Biosens Bioelectron 113:74–81
Ellman GL, Courtney KD, Andres VJ et al (1961) A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem Pharmacol 7:88–95
Baixauli F, Lopez-Otin C, Mittelbrunn M (2014) Exosomes and autophagy: coordinated mechanisms for the maintenance of cellular fitness. Front Immunol 5:403
Fader CM, Sanchez D, Furlan M et al (2008) Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic 9:230–250
Szatmari Z, Sass M (2014) The autophagic roles of Rab small GTPases and their upstream regulators: a review. Autophagy 10:1154–1166
Vanlandingham PA, Ceresa BP (2009) Rab7 regulates late endocytic trafficking downstream of multivesicular body biogenesis and cargo sequestration. J Biol Chem 284:12110–12124
Garcia NA, Ontoria-Oviedo I, Gonzalez-King H et al (2015) Glucose starvation in cardiomyocytes enhances exosome secretion and promotes angiogenesis in endothelial cells. PLoS ONE 10:e138849
Hernandez-Garcia MS, Miranda-Ozuna J, Salazar-Villatoro L et al (2019) Biogenesis of autophagosome in trichomonas vaginalis during macroautophagy induced by rapamycin-treatment and iron or glucose starvation conditions. J Eukaryot Microbiol. https://doi.org/10.1111/jeu.12712
Roa-Mansergas X, Fado R, Atari M et al (2018) CPT1C promotes human mesenchymal stem cells survival under glucose deprivation through the modulation of autophagy. Sci Rep 8:6997
Stein K, Winters C, Chiang HL (2017) Vps15p regulates the distribution of cup-shaped organelles containing the major eisosome protein Pil1p to the extracellular fraction required for endocytosis of extracellular vesicles carrying metabolic enzymes. Biol Cell 109:190–209
Abdulrahman BA, Abdelaziz DH, Schatzl HM (2018) Autophagy regulates exosomal release of prions in neuronal cells. J Biol Chem 293:8956–8968
Fader CM, Colombo MI (2006) Multivesicular bodies and autophagy in erythrocyte maturation. Autophagy 2:122–125
Rosenthal AK, Gohr CM, Mitton-Fitzgerald E et al (2015) Autophagy modulates articular cartilage vesicle formation in primary articular chondrocytes. J Biol Chem 290:13028–13038
Pietrocola F, Demont Y, Castoldi F et al (2017) Metabolic effects of fasting on human and mouse blood in vivo. Autophagy 13:567–578
Zhao SJ, Kong FQ, Cai W et al (2018) GIT1 contributes to autophagy in osteoclast through disruption of the binding of Beclin1 and Bcl2 under starvation condition. Cell Death Dis 9:1195
Gallagher LE, Williamson LE, Chan EY (2016) Advances in autophagy regulatory mechanisms. Cells 5:24
Schaaf MB, Houbaert D, Mece O et al (2019) Autophagy in endothelial cells and tumor angiogenesis. Cell Death Differ 26:665–679
Gomez-Sintes R, Villarejo-Zori B, Serrano-Puebla A et al (2017) Standard assays for the study of autophagy in the ex vivo retina. Cells 6:37
Xu J, Camfield R, Gorski SM (2018) The interplay between exosomes and autophagy—partners in crime. J Cell Sci 131:jcs215210
Desdin-Mico G, Mittelbrunn M (2017) Role of exosomes in the protection of cellular homeostasis. Cell Adhes Migr 11:127–134
Luiken JJ, Aerts JM, Meijer AJ (1996) The role of the intralysosomal pH in the control of autophagic proteolytic flux in rat hepatocytes. Eur J Biochem 235:564–573
Mauthe M, Orhon I, Rocchi C et al (2018) Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy 14:1435–1455
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (No. 81874146 to YY).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, X., Yin, X. & Yang, Y. Rasal2 suppresses breast cancer cell proliferation modulated by secretory autophagy. Mol Cell Biochem 462, 115–122 (2019). https://doi.org/10.1007/s11010-019-03615-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-019-03615-7