Abstract
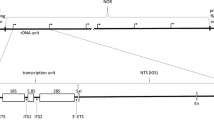
In human cells, the intergenic spacers (IGS), which separate ribosomal genes, are complex approximately 30 kb-long loci. Recent studies indicate that all, or almost all, parts of IGS may be transcribed, and that at least some of them are involved in the regulation of the ribosomal DNA (rDNA) transcription, maintenance of the nucleolar architecture, and response of the cell nucleus to stress. However, since each cell contains hundreds not quite identical copies of IGS, the structure and functions of this locus remain poorly understood, and the dynamics of its products has not been specially studied. In this work, we used quantitative PCR to measure the expression levels of various rDNA regions at different times after inhibition of the transcription by Actinomycin D applied in high doses. This approach allowed us to measure real or extrapolated half-life times of some IGS loci. Our study reveals characteristic dynamic patterns suggestive of various pathways of RNA utilization and decay.






Similar content being viewed by others
References
Agrawal S, Ganley AR (2015) Complete sequence construction of the highly repetitive ribosomal RNA gene repeats in eukaryotes using whole genome sequence data. Methods Mol Biol 1455:161–181
Agrawal S, Ganley ARD (2018) The conservation landscape of the human ribosomal RNA gene repeats. PLoS ONE 13(12):e0207531
Anosova I, Melnik S, Tripsianes K, Kateb F, Grummt I, Sattler M (2015) A novel RNA binding surface of the TAM domain of TIP5/BAZ2A mediates epigenetic regulation of rRNA genes. Nucleic Acids Res 43:5208–5220
Audas TE, Lee S (2016) Stressing out over long noncoding RNA. Biochim Biophys Acta 1859(1):184–191
Audas TE, Jacob MD, Lee S (2012a) The nucleolar detention pathway: a cellular strategy for regulating molecular networks. Cell Cycle 11:2059–2062
Audas TE, Jacob MD, Lee S (2012b) Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol Cell 45:147–157
Bensaude O (2011) Inhibiting eukaryotic transcription. Which compound to choose? How to evaluate its activity? Transcription 2(3):103–108
Bierhoff H, Dammert MA, Brocks D, Dambacher S, Schotta G, Grummt I (2014) Quiescence-induced LncRNAs trigger H4K20 trimethylation and transcriptional silencing. Mol Cell 54(4):675–682
Caburet S, Conti C, Schurra Lebofsky CR, Edelstein SJ, Bensimon A (2005) Human ribosomal RNA gene arrays display a broad range of palindromic structures. Genome Res 15:1079–1085
Caudron-Herger M, Pankert T, Seiler J, Németh A, Voit R, Grummt I, Rippe K (2015) Alu element-containing RNAs maintain nucleolar structure and function. EMBO J 34(22):2758–2774
Conconi A, Widmer RM, Koller T, Sogo JM (1989) Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell 57:753–761
Copenhaver GP, Putnam CD, Denton ML, Pikaard CS (1994) The RNA polymerase I transcription factor UBF is a sequence-tolerant HMG-box protein that can recognize structured nucleic acids. Nucleic Acids Res 22:2651–2657
Diermeier SD, Németh A, Rehli M, Grummt I, Längst G (2013) Chromatin-specific regulation of mammalian rDNA transcription by clustered TTF-I binding sites. PLoS Genet 9:e1003786
Eichler DC, Craig N (1994) Processing of eukaryotic ribosomal RNA. Prog Nucleic Acid Res Mol Biol 49:197–239
Floutsakou I, Agrawal S, Nguyen TT, Seoighe C, Ganley AR, McStay B (2013) The shared genomic architecture of human nucleolar organizer regions. Genome Res 23(12):2003–2012
Gonzalez IL, Sylvester JE (1995) Complete sequence of the 43-kb human ribosomal DNA repeat: analysis of the intergenic spacer. Genomics 27:320–328
Gonzalez IL, Sylvester JE (2001) Human rDNA: evolutionary patterns within the genes and tandem arrays derived from multiple chromosomes. Genomics 73:255–263
Gonzalez IL, Wu S, Li W-M, Kuo BA, Sylvester JE (1992) Human ribosomal RNA intergenic spacer sequence. Nucleic Acids Res 20:5846
Grummt I, Pikaard CS (2003) Epigenetic silencing of RNA polymerase I transcription. Nat Rev Mol Cell Biol 4:641–649
Hadjiolova KV, Hadjiolov AA, Bachellerie JP (1995) Actinomycin D stimulates the transcription of rRNA minigenes transfected into mouse cells. Implications for the in vivo hypersensitivity of rRNA gene transcription. Eur J Biochem 228:605–615
Haltiner MM, Smale ST, Tjian R (1986) Two distinct promoter elements in the human rRNA gene identified by linker scanning mutagenesis. Mol Cell Biol 6(1):227–235
Henderson AS, Warburton D, Atwood KC (1972) Location of ribosomal DNA in the human chromosome complement. Proc Natl Acad Sci USA 69:3394–3398
Hornáček M, Kováčik L, Mazel T, Cmarko D, Bártová E, Raška I, Smirnov E (2017) Fluctuations of pol I and fibrillarin contents of the nucleoli. Nucleus 8(4):421–432
Hu CH, McStay B, Jeong SW, Reeder RH (1994) xUBF, an RNA polymerase I transcription factor, binds crossover DNA with low sequence specificity. Mol Cell Biol 14:2871–2882
Jacob MD, Audas TE, Mullineux ST, Lee S (2012) Where no RNA polymerase has gone before: novel functional transcripts derived from the ribosomal intergenic spacer. Nucleus 3:315–319
Lam YW, Trinkle-Mulcahy L (2015) New insights into nucleolar structure and function. F1000Prime Rep 7:48
Lebofsky R, Bensimon A (2005) DNA replication origin plasticity and perturbed fork progression in human inverted repeats. Mol Cell Biol 25:6789–6797
Long EO, Dawid IB (1980) Repeated genes in eukaryotes. Annu Rev Biochem 49:727–764
Maden BE, Dent CL, Farrell TE, Garde J, McCallum FS, Wakeman JA (1987) Clones of human ribosomal DNA containing the complete 18 S-rRNA and 28 S-rRNA genes. Characterization, a detailed map of the human ribosomal transcription unit and diversity among clones. Biochem J 246:519–527
Mayer C, Schmitz KM, Li J, Grummt I, Santoro R (2006) Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol Cell 22:351–361
Mayer C, Neubert M, Grummt I (2008) The structure of NoRC-associated RNA is crucial for targeting the chromatin remodelling complex NoRC to the nucleolus. EMBO Rep 9(8):774–780
McStay B, Grummt I (2008) The epigenetics of rRNA genes: from molecular to chromosome biology. Annu Rev Cell Dev Biol 24:131–157
Moss T, Langlois F, Gagnon-Kugler T, Stefanovsky V (2007) A housekeeper with power of attorney: the rRNA genes in ribosome biogenesis. Cell Mol Life Sci 64:29–49
Moss T, Mars JC, Tremblay MG, Sabourin-Felix M (2019) The chromatin landscape of the ribosomal RNA genes in mouse and human. Chromosome Res 27(1–2):31–40
Nazar RN (2004) Ribosomal RNA processing and ribosome biogenesis in eukaryotes. IUBMB Life 56:457–465
Németh A, Grummt I (2018) Dynamic regulation of nucleolar architecture. Curr Opin Cell Biol 52:105–111
Popov A, Smirnov E, Kováčik L, Raška O, Hagen G, Stixová L, Raška I (2013) Duration of the first steps of the human rRNA processing. Nucleus 4(2):134–141
Santoro R (2014) Analysis of chromatin composition of repetitive sequences: the ChIP-Chop assay. Methods Mol Biol 1094:319–328
Santoro R, De Lucia F (2005) Many players, one goal: how chromatin states are inherited during cell division. Biochem Cell Biol 83:332–343
Santoro R, Schmitz KM, Sandoval J, Grummt I (2010) Intergenic transcripts originating from a subclass of ribosomal DNA repeats silence ribosomal RNA genes in trans. EMBO Rep 11(1):52–58
Sasaki T, Okazaki T, Muramatsu M, Kominami R (1987) Variation among mouse ribosomal RNA genes within and between chromosomes. Mol Biol Evol 4:594–601
Shcherbik N, Wang M, Lapik YR, Srivastava L, Pestov DG (2010) Polyadenylation and degradation of incomplete RNA polymerase I transcripts in mammalian cells. EMBO Rep 11:106–111
Smirnov E, Cmarko D, Mazel T, Hornáček M, Raška I (2016) Nucleolar DNA: the host and the guests. Histochem Cell Biol 145(4):359–372
Sylvester JE, Gonzalez IL, Mougey EB (2003) In: Olson M (ed) The nucleolus. Landes Bioscience, Austin, pp 58–73
Warner JR (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 24:437–440
Wehner S, Dörrich AK, Ciba P, Wilde A, Marz M (2014) pRNA. NoRC-associated RNA of rRNA operons. RNA Biol 11(1):3–9
Zentner GE, Saiakhova A, Manaenkov P, Adams MD, Scacheri PC (2011) Integrative genomic analysis of human ribosomal DNA. Nucleic Acids Res 39:4949–4960
Zentner GE, Balow SA, Scacheri PC (2014) Genomic characterization of the mouse ribosomal DNA locus. G3 G3(4):243–254
Zhao Z, Dammert MA, Grummt I, Bierhoff H (2016) lncRNA-induced nucleosome repositioning reinforces transcriptional repression of rRNA genes upon hypotonic stress. Cell Rep 14(8):1876–1882
Zhao Z, Sentürk N, Song C, Grummt I (2018) lncRNA PAPAS tethered to the rDNA enhancer recruits hypophosphorylated CHD4/NuRD to repress rRNA synthesis at elevated temperatures. Genes Dev 32(11–12):836–848
Zillner K, Komatsu J, Filarsky K, Kalepu R, Bensimon A, Nemeth A (2015) Active human nucleolar organizer regions are interspersed with inactive rDNA repeats in normal and tumor cells. Epigenomics 7:363–378
Acknowledgements
This work was supported by the Grant Agency of Czech Republic (P302/12/G157, 19-21715S, and 19-19779S) and Charles University (Progres Q28).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Vacík, T., Kereïche, S., Raška, I. et al. Life time of some RNA products of rDNA intergenic spacer in HeLa cells. Histochem Cell Biol 152, 271–280 (2019). https://doi.org/10.1007/s00418-019-01804-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00418-019-01804-5