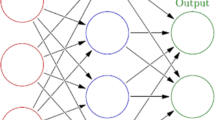
Abstract
Background
Metabolomics data, with its complex covariance structure, is typically modelled by projection-based machine learning (ML) methods such as partial least squares (PLS) regression, which project data into a latent structure. Biological data are often non-linear, so it is reasonable to hypothesize that metabolomics data may also have a non-linear latent structure, which in turn would be best modelled using non-linear equations. A non-linear ML method with a similar projection equation structure to PLS is artificial neural networks (ANNs). While ANNs were first applied to metabolic profiling data in the 1990s, the lack of community acceptance combined with limitations in computational capacity and the lack of volume of data for robust non-linear model optimisation inhibited their widespread use. Due to recent advances in computational power, modelling improvements, community acceptance, and the more demanding needs for data science, ANNs have made a recent resurgence in interest across research communities, including a small yet growing usage in metabolomics. As metabolomics experiments become more complex and start to be integrated with other omics data, there is potential for ANNs to become a viable alternative to linear projection methods.
Aim of review
We aim to first describe ANNs and their structural equivalence to linear projection-based methods, including PLS regression. We then review the historical, current, and future uses of ANNs in the field of metabolomics.
Key scientific concept of review
Is metabolomics ready for the return of artificial neural networks?





Similar content being viewed by others
References
Adadi, A., & Berrada, M. (2018). Peeking inside the black-box: A survey on Explainable Artificial Intelligence (XAI). IEEE Access, 6, 52138–52160.
AlaKorpela, M., Changani, K. K., Hiltunen, Y., Bell, J. D., Fuller, B. J., Bryant, D. J., et al. (1997). Assessment of quantitative artificial neural network analysis in a metabolically dynamic ex vivo P-31 NMR pig liver study. Magnetic Resonance in Medicine, 38, 840–844.
Alakwaa, F. M., Chaudhary, K., & Garmire, L. X. (2018). Deep learning accurately predicts estrogen receptor status in breast cancer metabolomics data. Journal of Proteome Research, 17, 337–347.
Aliakbarzadeh, G., Sereshti, H., & Parastar, H. (2016). Pattern recognition analysis of chromatographic fingerprints of Crocus sativus L. secondary metabolites towards source identification and quality control. Analytical and Bioanalytical Chemistry, 408, 3295–3307.
Allen, F., Pon, A., Greiner, R., & Wishart, D. (2016). Computational prediction of electron ionization mass spectra to assist in GC/MS compound identification. Analytical Chemistry, 88, 7689–7697.
Alom, M. Z., Taha, T. M., Yakopcic, C., Westberg, S., Sidike, P., Nasrin, M. S., et al. (2018). The history began from AlexNet: A comprehensive survey on deep learning approaches. arXiv:1803.01164.
Anthony, M. L., Rose, V. S., Nicholson, J. K., & Lindon, J. C. (1995). Classification of toxin-induced changes in 1H NMR spectra of urine using an artificial neural network. Journal of Pharmaceutical and Biomedical Analysis, 13, 205–211.
Asakura, T., Date, Y., & Kikuchi, J. (2018). Application of ensemble deep neural network to metabolomics studies. Analytica Chimica Acta, 1037, 230–236.
Azmi, J., Griffin, J. L., Antti, H., Shore, R. F., Johansson, E., Nicholson, J. K., et al. (2002). Metabolic trajectory characterisation of xenobiotic-induced hepatotoxic lesions using statistical batch processing of NMR data. Analyst, 127, 271–276.
Banerjee, P., Barman, S. R., Sikdar, D., Roy, U., Mukhopadhayay, A., & Das, P. (2017). Enhanced degradation of ternary dye effluent by developed bacterial consortium with RSM optimization, ANN modeling and toxicity evaluation. Desalination and Water Treatment, 72, 249–265.
Barnette, D. A., Davis, M. A., Dang, N. L., Pidugu, A. S., Hughes, T., Swamidass, S. J., et al. (2018). Lamisil (terbinafine) toxicity: Determining pathways to bioactivation through computational and experimental approaches. Biochemical Pharmacology, 156, 10–21.
Basheer, I. A., & Hajmeer, M. (2000). Artificial neural networks: Fundamentals, computing, design, and application. Journal of Microbiological Methods, 43, 3–31.
Beger, R. D., Dunn, W. B., Bandukwala, A., Bethan, B., Broadhurst, D., Clish, C. B., et al. (2019). Towards quality assurance and quality control in untargeted metabolomics studies. Metabolomics, 15, 4.
Bengio, Y., Ducharme, R., Vincent, P., & Jauvin, C. (2003). A neural probabilistic language model. Journal of Machine Learning Research, 3, 1137–1155.
Bica, I., Velickovic, P., Xiao, H., & Li, P. (2018). Multi-omics data integration using cross-modal neural networks. In European symposium on artificial neural networks, computational intelligence and machine learning (pp. 385–390).
Bostrom, N., & Yudkowsky, E. (2014). Chapter 15—The ethics of artificial intelligence. The Cambridge handbook of artificial intelligence. Cambridge: Cambridge University Press.
Breiman, L. (2001). Statistical modeling: The two cultures. Statistical Science, 16, 199–231.
Broadhurst, D. (2019). Is metabolomics ready for the return of artificial neural networks? Retrieved August 25, 2019, from https://doi.org/10.6084/m9.figshare.8326529.v1
Broadhurst, D., Goodacre, R., Reinke, S. N., Kuligowski, J., Wilson, I. D., Lewis, M. R., et al. (2018). Guidelines and considerations for the use of system suitability and quality control samples in mass spectrometry assays applied in untargeted clinical metabolomic studies. Metabolomics, 14, 72.
Broadhurst, D. I., & Kell, D. B. (2006). Statistical strategies for avoiding false discoveries in metabolomics and related experiments. Metabolomics, 2, 171–196.
Cambria, E., & White, B. (2014). Jumping NLP curves: A review of natural language processing research. IEEE Computational Intelligence Magazine, 9, 48–57.
Cavill, R., Jennen, D., Kleinjans, J., & Briede, J. J. (2016). Transcriptomic and metabolomic data integration. Briefings in Bioinformatics, 17, 891–901.
Chagas-Paula, D. A., Oliveira, T. B., Zhang, T., Edrada-Ebel, R., & Da Costa, F. B. (2015). Prediction of anti-inflammatory plants and discovery of their biomarkers by machine learning algorithms and metabolomic studies. Planta Medica, 81, 450–458.
Chaudhary, K., Poirion, O. B., Lu, L., & Garmire, L. X. (2018). Deep learning-based multi-omics integration robustly predicts survival in liver cancer. Clinical Cancer Research, 24, 1248–1259.
Chen, J. X. (2016). The evolution of computing: AlphaGo. Computing in Science & Engineering, 18, 4.
Chollet, F. (2015). Keras. Retrieved August 27, 2019, from https://keras.io/
Chollet, F. (2018). Chapter 2: Before we begin: The mathematical building blocks of neural networks, deep learning with Python. New York: Manning Publications Co.
Chung, N. C., Mirza, B., Choi, H., Wang, J., Wang, D., Ping, P., et al. (2019). Unsupervised classification of multi-omics data during cardiac remodeling using deep learning. Methods, 166, 66–73.
Cireşan, D. C., Giusti, A., Gambardella, L. M., & Schmidhuber, J. (2012a). Deep neural networks segment neuronal membranes in electron microscopy images. In Proceedings of the 25th international conference on neural information processing systems (Vol. 1, pp. 2843–2851).
Cireşan, D. C., Giusti, A., Gambardella, L. M., & Schmidhuber, J. (2013). Mitosis detection in breast cancer histology images with deep neural networks. Medical Image Computing and Computer-Assisted Intervention, 1, 411–418.
Cireşan, D. C., Meier, U., Masci, J., & Schmidhuber, J. (2012b). Multi-column deep neural network for traffic sign classification. Neural Networks, 32, 333–338.
Cortina, P. R., Santiago, A. N., Sance, M. M., Peralta, I. E., Carrari, F., & Asis, R. (2018). Neuronal network analyses reveal novel associations between volatile organic compounds and sensory properties of tomato fruits. Metabolomics, 14, 15.
Date, Y., & Kikuchi, J. (2018). Application of a deep neural network to metabolomics studies and its performance in determining important variables. Analytical Chemistry, 90, 1805–1810.
Deelen, J., Kettunen, J., Fischer, K., van der Spek, A., Trompet, S., Kastenmüller, G., et al. (2019). A metabolic profile of all-cause mortality risk identified in an observational study of 44,168 individuals. Nature Communications, 10, 3346.
Dong, W. J., Zhao, J. P., Hu, R. S., Dong, Y. P., & Tan, L. H. (2017). Differentiation of Chinese robusta coffees according to species, using a combined electronic nose and tongue, with the aid of chemometrics. Food Chemistry, 229, 743–751.
Dunn, W. B., Broadhurst, D. I., Atherton, H. J., Goodacre, R., & Griffin, J. L. (2011). Systems level studies of mammalian metabolomes: The roles of mass spectrometry and nuclear magnetic resonance spectroscopy. Chemical Society Reviews, 40, 387–426.
Dunn, W. B., Lin, W., Broadhurst, D., Begley, P., Brown, M., Zelena, E., et al. (2015). Molecular phenotyping of a UK population: Defining the human serum metabolome. Metabolomics, 11, 9–26.
Egmont-Petersen, M., de Ridder, D., & Handels, H. (2002). Image processing with neural networks—A review. Pattern Recognition, 35, 2279–2301.
Eraslan, G., Avsec, Ž., Gagneur, J., & Theis, F. J. (2019). Deep learning: New computational modelling techniques for genomics. Nature Reviews Genetics, 20, 389–403.
Erevelles, S., Fukawa, N., & Swayne, L. (2016). Big data consumer analytics and the transformation of marketing. Journal of Business Research, 69, 897–904.
Falcini, F., Lami, G., & Costanza, A. M. (2017). Deep learning in automotive software. IEEE Software, 34, 56–63.
Fatemi, M. H., Shahroudi, E. M., & Amini, Z. (2015). Development of quantitative interspecies toxicity relationship modeling of chemicals to fish. Journal of Theoretical Biology, 380, 16–23.
Fayolle, P., Picque, D., & Corrieu, G. (1997). Monitoring of fermentation processes producing lactic acid bacteria by mid-infrared spectroscopy. Vibrational Spectroscopy, 14, 247–252.
Ferrucci, D. A. (2012). Introduction to “This Is Watson”. IBM Journal of Research and Development, 56, 235–249.
Flagel, L., Brandvain, Y., & Schrider, D. R. (2018). The unreasonable effectiveness of convolutional neural networks in population genetic inference. Molecular Biology and Evolution, 36, 220–238.
Francescatto, M., Chierici, M., Rezvan Dezfooli, S., Zandonà, A., Jurman, G., & Furlanello, C. (2018). Multi-omics integration for neuroblastoma clinical endpoint prediction. Biology Direct, 13, 5.
Frisvad, J. C. (1992). Chemometrics and chemotaxonomy: A comparison of multivariate statistical methods for the evaluation of binary fungal secondary metabolite data. Chemometrics and Intelligent Laboratory Systems, 14, 253–269.
Fukushima, K. (1980). Neocognitron: A self-organizing neural network model for a mechanism of pattern recognition unaffected by shift in position. Biological Cybernetics, 36, 193–202.
Gardner, M. W., & Dorling, S. R. (1998). Artificial neural networks (the multilayer perceptron)—A review of applications in the atmospheric sciences. Atmospheric Environment, 32, 2627–2636.
Garson, G. D. (1991). Interpreting neural network connection weights. AI Expert, 6, 47–51.
Geladi, P., & Kowalski, B. R. (1986). Partial least-squares regression: A tutorial. Analytica Chimica Acta, 185, 1–17.
Goodacre, R. (2003). Explanatory analysis of spectroscopic data using machine learning of simple, interpretable rules. Vibrational Spectroscopy, 32, 33–45.
Goodacre, R., & Kell, D. B. (1993). Rapid and quantitative analysis and bioprocesses using pyrolysis mass spectrometry and neural networks: Application to indole production. Analytica Chimica Acta, 279, 17–26.
Goodacre, R., & Kell, D. B. (1996). Correction of mass spectral drift using artificial neural networks. Analytical Chemistry, 68, 271–280.
Goodacre, R., Kell, D. B., & Bianchi, G. (1992). Neural networks and olive oil. Nature, 359, 594.
Goodacre, R., Kell, D. B., & Bianchi, G. (1993). Rapid assessment of the adulteration of virgin olive oils by other seed oils using pyrolysis mass spectrometry and artificial neural networks. Journal of the Science of Food and Agriculture, 63, 297–307.
Goodacre, R., Rischert, D. J., Evans, P. M., & Kell, D. B. (1996a). Rapid authentication of animal cell lines using pyrolysis mass spectrometry and auto-associative artificial neural networks. Cytotechnology, 21, 231–241.
Goodacre, R., Rooney, P. J., & Kell, D. B. (1998). Discrimination between methicillin-resistant and methicillin-susceptible Staphylococcus aureus using pyrolysis mass spectrometry and artificial neural networks. Journal of Antimicrobial Chemotherapy, 41, 27–34.
Goodacre, R., Timmins, É. M., Jones, A., Kell, D. B., Maddock, J., Heginbothom, M. L., et al. (1997). On mass spectrometer instrument standardization and interlaboratory calibration transfer using neural networks. Analytica Chimica Acta, 348, 511–532.
Goodacre, R., Timmins, E. M., Rooney, P. J., Rowland, J. J., & Kell, D. B. (1996b). Rapid identification of Streptococcus and Enterococcus species using diffuse reflectance-absorbance Fourier transform infrared spectroscopy and artificial neural networks. FEMS Microbiology Letters, 140, 233–239.
Goodacre, R., Trew, S., Wrigleyjones, C., Neal, M. J., Maddock, J., Ottley, T. W., et al. (1994). Rapid screening for metabolite overproduction in fermentor broths, using pyrolysis mass-spectrometry with multivariate calibration and artificial neural networks. Biotechnology and Bioengineering, 44, 1205–1216.
Goodfellow, I., Bengio, Y., & Courville, A. (2016). Deep learning. Massachusetts, United States of America: MIT press.
Grapov, D., Fahrmann, J., Wanichthanarak, K., & Khoomrung, S. (2018). Rise of deep learning for genomic, proteomic, and metabolomic data integration in precision medicine. Omics-a Journal of Integrative Biology, 22, 630–636.
Gromski, P. S., Muhamadali, H., Ellis, D. I., Xu, Y., Correa, E., Turner, M. L., et al. (2015). A tutorial review: Metabolomics and partial least squares-discriminant analysis—A marriage of convenience or a shotgun wedding. Analytica Chimica Acta, 879, 10–23.
Guo, J. R., Chen, Q. Q., Lam, C. W. K., Wang, C. Y., Wong, V. K. W., Xu, F. G., et al. (2015). Application of artificial neural network to investigate the effects of 5-fluorouracil on ribonucleotides and deoxyribonucleotides in HepG2 cells. Scientific Reports, 5, 14.
Guo, Y., Liu, Y., Oerlemans, A., Lao, S., Wu, S., & Lew, M. S. (2016). Deep learning for visual understanding: A review. Neurocomputing, 187, 27–48.
Hall, L. M., Hill, D. W., Bugden, K., Cawley, S., Hall, L. H., Chen, M. H., et al. (2018). Development of a reverse phase HPLC Retention Index Model for nontargeted metabolomics using synthetic compounds. Journal of Chemical Information and Modeling, 58, 591–604.
Hall, L. M., Hill, D. W., Menikarachchi, L. C., Chen, M. H., Hall, L. H., & Grant, D. F. (2015). Optimizing artificial neural network models for metabolomics and systems biology: An example using HPLC retention index data. Bioanalysis, 7, 939–955.
Hamid, J. S., Hu, P., Roslin, N. M., Ling, V., Greenwood, C. M. T., & Beyene, J. (2009). Data integration in genetics and genomics: Methods and challenges. Human Genomics and Proteomics, 2009, 869093.
Harthun, S., Matischak, K., & Friedl, P. (1998). Simultaneous prediction of human antithrombin III and main metabolites in animal cell culture processes by near-infrared spectroscopy. Biotechnology Techniques, 12, 393–397.
Hettinga, K. A., de Bok, F. A. M., & Lam, T. (2015). Short communication: Practical issues in implementing volatile metabolite analysis for identifying mastitis pathogens. Journal of Dairy Science, 98, 7906–7910.
Holmes, E., Loo, R. L., Stamler, J., Bictash, M., Yap, I. K., Chan, Q., et al. (2008). Human metabolic phenotype diversity and its association with diet and blood pressure. Nature, 453, 396–400.
Holzinger, A., Biemann, C., Pattichis, C. S., & Kell, D. B. (2017). What do we need to build explainable AI systems for the medical domain? arXiv:1712.09923.
Hotelling, H. (1933). Analysis of a complex of statistical variables into principal components. Journal of Educational Psychology, 24, 417–441.
Huang, T., Lan, L., Fang, X. X., An, P., Min, J. X., & Wang, F. D. (2015). Promises and challenges of big data computing in health sciences. Big Data Research, 2, 2–11.
Huang, Z., Zhan, X., Xiang, S., Johnson, T. S., Helm, B., Yu, C. Y., et al. (2019). SALMON: Survival analysis learning with multi-omics neural networks on breast cancer. Frontiers in Genetics, 10, 166.
Hughes, T. B., & Swamidass, S. J. (2017). Deep learning to predict the formation of quinone species in drug metabolism. Chemical Research in Toxicology, 30, 642–656.
Inglese, P., McKenzie, J. S., Mroz, A., Kinross, J., Veselkov, K., Holmes, E., et al. (2017). Deep learning and 3D-DESI imaging reveal the hidden metabolic heterogeneity of cancer. Chemical Science, 8, 3500–3511.
Jolliffe, I. T. (1982). A note on the use of principal components in regression. Journal of the Royal Statistical Society: Series C (Applied Statistics), 31, 300–303.
Jolliffe, I. T. (2002). Principal component analysis (2nd ed.). New York: Springer.
Kaartinen, J., Mierisova, S., Oja, J. M. E., Usenius, J. P., Kauppinen, R. A., & Hiltunen, Y. (1998). Automated quantification of human brain metabolites by artificial neural network analysis from in vivo single-voxel H-1 NMR spectra. Journal of Magnetic Resonance, 134, 176–179.
Kendall, M. G. (1957). A course in multivariate analysis. New York: Hafner Publishing Company.
Krizhevsky, A., Sutskever, I., & Hinton, G. E. (2012). ImageNet classification with deep convolutional neural networks. In Proceedings of the 25th international conference on neural information processing systems (Vol. 1, pp. 1097–1105)
Lang, N. P., Butler, M. A., Massengill, J., Lawson, M., Stotts, R. C., Hauerjensen, M., et al. (1994). Rapid metabolic phenotypes for acetyltransferase and cytochrome P4501A2 and putative exposure to food-borne heterocyclic amines increase the risk for colorectal cancer or polyps. Cancer Epidemiology, Biomarkers and Prevention, 3, 675–682.
Lenz, I., Lee, H., & Saxena, A. (2015). Deep learning for detecting robotic grasps. The International Journal of Robotics Research, 34, 705–724.
Levine, S., Pastor, P., Krizhevsky, A., Ibarz, J., & Quillen, D. (2018). Learning hand-eye coordination for robotic grasping with deep learning and large-scale data collection. The International Journal of Robotics Research, 37, 421–436.
Lindon, J. C., Nicholson, J. K., Holmes, E., Antti, H., Bollard, M. E., Keun, H., et al. (2003). Contemporary issues in toxicology the role of metabonomics in toxicology and its evaluation by the COMET project. Toxicology and Applied Pharmacology, 187, 137–146.
Löfstedt, T., & Trygg, J. (2011). OnPLS—A novel multiblock method for the modelling of predictive and orthogonal variation. Journal of Chemometrics, 25, 441–455.
Long, N. P., Lim, D. K., Mo, C., Kim, G., & Kwon, S. W. (2017). Development and assessment of a lysophospholipid-based deep learning model to discriminate geographical origins of white rice. Scientific Reports, 7, 10.
Luckow, A., Cook, M., Ashcraft, N., Weill, E., Djerekarov, E., & Vorster, B. (2016). Deep learning in the automotive industry: Applications and tools. IEEE International Conference on Big Data, 1, 3759–3768.
Manning, C., Surdeanu, M., Bauer, J., Finkel, J., Bethard, S., & McClosky, D. (2014). The Stanford CoreNLP natural language processing toolkit. In Proceedings of 52nd annual meeting of the association for computational linguistics: System demonstrations (Vol. 1, pp. 55–60).
Maudsley, S., Devanarayan, V., Martin, B., Geerts, H., & Brain Health Modeling Initiative. (2018). Intelligent and effective informatic deconvolution of “Big Data” and its future impact on the quantitative nature of neurodegenerative disease therapy. Alzheimers & Dementia, 14, 961–975.
McCulloch, W. S., & Pitts, W. (1943). A logical calculus of the ideas immanent in nervous activity. The Bulletin of Mathematical Biophysics, 5, 115–133.
Moen, B. E., Nilsson, R., Nordlinder, R., Ovrebo, S., Bleie, K., Skorve, A. H., et al. (1996). Assessment of exposure to polycyclic aromatic hydrocarbons in engine rooms by measurement of urinary 1-hydroxypyrene. Occupational and Environmental Medicine, 53, 692–696.
Moore, G. E. (1975). Progress in digital integrated electronics. Electron Devices Meeting, 21, 11–13.
Morin, F., & Bengio, Y. (2005). Hierarchical probabilistic neural network language model. International Conference on Artificial Intelligence and Statistics, 5, 246–252.
Mosconi, F., Julou, T., Desprat, N., Sinha, D. K., Allemand, J.-F., Croquette, V., et al. (2008). Some nonlinear challenges in biology. Nonlinearity, 21, 131–147.
Oh, K.-S., & Jung, K. (2004). GPU implementation of neural networks. Pattern Recognition, 37, 1311–1314.
Olden, J. D., & Jackson, D. A. (2002). Illuminating the “black box”: A randomization approach for understanding variable contributions in artificial neural networks. Ecological Modelling, 154, 135–150.
Olden, J. D., Joy, M. K., & Death, R. G. (2004). An accurate comparison of methods for quantifying variable importance in artificial neural networks using simulated data. Ecological Modelling, 178, 389–397.
Ou, X. Q., Li, H., Yang, X. M., Tan, M. L., Ao, H., & Wang, J. (2015). Artificial neural network analysis of Xinhui pericarpium citri reticulatae using gas chromatography—Mass spectrometer—Automated mass spectral deconvolution and identification system. Tropical Journal of Pharmaceutical Research, 14, 2071–2075.
Pecnik, K., Todorovic, V., Bosnjak, M., Cemazar, M., Kononenko, I., Sersa, G., et al. (2018). The general explanation method with NMR spectroscopy enables the identification of metabolite profiles specific for normal and tumor cell lines. ChemBioChem, 19, 2066–2071.
Pérez-Enciso, M., & Tenenhaus, M. (2003). Prediction of clinical outcome with microarray data: A partial least squares discriminant analysis (PLS-DA) approach. Human Genetics, 112, 581–592.
Peters, W., Gang, E. S., Okazaki, H., Solingen, S., Kobayashi, Y., Karagueuzian, H. S., et al. (1991). Acute effects of intravenous propafenone on the internal ventricular defibrillation threshold in the anesthetized dog. American Heart Journal, 122, 1355–1360.
Pinu, R. F., Beale, J. D., Paten, M. A., Kouremenos, K., Swarup, S., Schirra, J. H., et al. (2019). Systems biology and multi-omics integration: Viewpoints from the metabolomics research community. Metabolites, 9, 76.
Qiu, S., Yang, W. Z., Yao, C. L., Qiu, Z. D., Shi, X. J., Zhang, J. X., et al. (2016). Nontargeted metabolomic analysis and “commercial-homophyletic” comparison-induced biomarkers verification for the systematic chemical differentiation of five different parts of Panax ginseng. Journal of Chromatography A, 1453, 78–87.
Rawat, W., & Wang, Z. (2017). Deep convolutional neural networks for image classification: A comprehensive review. Neural Computation, 29, 2352–2449.
Reinke, S. N., Galindo-Prieto, B., Skotare, T., Broadhurst, D. I., Singhania, A., Horowitz, D., et al. (2018). OnPLS-based multi-block data integration: A multivariate approach to interrogating biological interactions in asthma. Analytical Chemistry, 90, 13400–13408.
Risum, A. B., & Bro, R. (2019). Using deep learning to evaluate peaks in chromatographic data. Talanta, 204, 255–260.
Robinson, D. A. (1992). Implications of neural networks for how we think about brain function. Behavioral and Brain Sciences, 15, 644–655.
Rohart, F., Gautier, B., Singh, A., & Lê Cao, K.-A. (2017). mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Computational Biology, 13, e1005752.
Roundtable on Translating Genomic-Based Research for Health, Board on Health Sciences Policy, Health and Medicine Division and National Academies of Sciences, E., and Medicine. (2016). F, Large Genetic Cohort Studies: A Background in Siobhan, A., Steve, O. and Sarah, H.B. (Eds), Applying an implementation science approach to genomic medicine: Workshop summary. Washington, DC: The National Academies Press
Russakovsky, O., Deng, J., Su, H., Krause, J., Satheesh, S., Ma, S., et al. (2015). ImageNet large scale visual recognition challenge. International Journal of Computer Vision, 115, 211–252.
Russell, S., Hauert, S., Altman, R., & Veloso, M. (2015). Ethics of artificial intelligence. Nature, 521, 415–416.
Samaraweera, M. A., Hall, L. M., Hill, D. W., & Grant, D. F. (2018). Evaluation of an artificial neural network retention index model for chemical structure identification in nontargeted metabolomics. Analytical Chemistry, 90, 12752–12760.
Samek, W., Montavon, G., Vedaldi, A., Hansen, L. K., & Muller, K.-R. (Eds.). (2019). Explainable AI: Interpreting, explaining and visualizing deep learning. Basel: Springer International Publishing.
Samek, W., Wiegand, T., & Müller, K.-R. (2017). Explainable artificial intelligence: Understanding, visualizing and interpreting deep learning models. arXiv:1708.08296.
Saunders, G., Baudis, M., Becker, R., Beltran, S., Béroud, C., Birney, E., et al. (2019) Leveraging European infrastructures to access 1 million human genomes by 2022. Nature Reviews Genetics 1–9.
Schalkoff, R. J. (1997). Artificial neural networks, (International ed.). London: McGraw-Hill.
Schmidhuber, J. (2012) Multi-column deep neural networks for image classification. In Proceedings of the 2012 IEEE conference on computer vision and pattern recognition (CVPR) (pp. 3642–3649).
Seber, G. A. F. (2004). Multivariate observations (2nd ed.). New Jersey: Wiley.
Sharifi-Noghabi, H., Zolotareva, O., Collins, C. C., & Ester, M. (2019). MOLI: Multi-omics late integration with deep neural networks for drug response prediction. Bioinformatics, 35, i501–i509.
Shen, D., Wu, G., & Suk, H.-I. (2017). Deep learning in medical image analysis. Annual Review of Biomedical Engineering, 19, 221–248.
Simard, P. Y., Steinkraus, D., & Platt, J. C. (2003). Best practices for convolutional neural networks applied to visual document analysis. International Conference on Document Analysis and Recognition, 3, 1–6.
Sjogren, M., Ehrenberg, L., & Rannug, U. (1996). Relevance of different biological assays in assessing initiating and promoting properties of polycyclic aromatic hydrocarbons with respect to carcinogenic potency. Mutation Research-Fundamental and Molecular Mechanisms of Mutagenesis, 358, 97–112.
Tautenhahn, R., Patti, G. J., Rinehart, D., & Siuzdak, G. (2012). XCMS online: A web-based platform to process untargeted metabolomic data. Analytical Chemistry, 84, 5035–5039.
Trainor, P. J., DeFilippis, A. P., & Rai, S. N. (2017). Evaluation of classifier performance for multiclass phenotype discrimination in untargeted metabolomics. Metabolites, 7, 20.
Trivedi, D. K., Hollywood, K. A., & Goodacre, R. (2017). Metabolomics for the masses: The future of metabolomics in a personalized world. New Horizons in Translational Medicine, 3, 294–305.
Usenius, J. P., Tuohimetsa, S., Vainio, P., AlaKorpela, M., Hiltunen, Y., & Kauppinen, R. A. (1996). Automated classification of human brain tumours by neural network analysis using in vivo H-1 magnetic resonance spectroscopic metabolite phenotypes. NeuroReport, 7, 1597–1600.
Wang, F., Wang, B., Wang, L., Xiong, Z. Y., Gao, W., Li, P., et al. (2017). Discovery of discriminatory quality control markers for Chinese herbal medicines and related processed products by combination of chromatographic analysis and chemometrics methods: Radix Scutellariae as a case study. Journal of Pharmaceutical and Biomedical Analysis, 138, 70–79.
Wang, F.-Y., Zhang, J. J., Zheng, X., Wang, X., Yuan, Y., Dai, X., et al. (2016). Where does AlphaGo go: From church-turing thesis to AlphaGo thesis and beyond. IEEE/CAA Journal of Automatica Sinica, 3, 113–120.
Wold, H. (1975). Path models with latent variables: The NIPALS approach. Quantitative sociology (pp. 307–357). New York: Elsevier.
Wold, S., Esbensen, K., & Geladi, P. (1987). Principal component analysis. Chemometrics and Intelligent Laboratory Systems, 2, 37–52.
Wold, S., Johansson, E., & Cocchi, M. (1993). PLS: Partial least squares projections to latent structures, 3D QSAR in drug design: Theory methods and applications. Dordrecht: Kluwer/Escom.
Woldegebriel, M., & Derks, E. (2017). Artificial neural network for probabilistic feature recognition in liquid chromatography coupled to high-resolution mass spectrometry. Analytical Chemistry, 89, 1212–1221.
Wolff, M. S., Toniolo, P. G., Lee, E. W., Rivera, M., & Dubin, N. (1993). Blood levels of organochlorine residues and risk of breast cancer. Journal of the National Cancer Institute, 85, 648–652.
Worley, B., & Powers, R. (2013). Multivariate analysis in metabolomics. Current metabolomics, 1, 92–107.
Wu, T., Liu, S., Zhang, J., & Xiang, Y. (2017). Twitter spam detection based on deep learning. Proceedings of the Australasian Computer Science Week Multiconference, 1, 3.
Young, T., Hazarika, D., Poria, S., & Cambria, E. (2018). Recent trends in deep learning based natural language processing. IEEE Computational Intelligence Magazine, 13, 55–75.
Yue, T., & Wang, H. (2018) Deep learning for genomics: A concise overview. arXiv:1802.00810.
Zhang, Z., Beck, M. W., Winkler, D. A., Huang, B., Sibanda, W., Goyal, H., et al. (2018). Opening the black box of neural networks: Methods for interpreting neural network models in clinical applications. Annals of Translational Medicine, 6, 216.
Zhang, X. X., Li, Y. Z., Liang, Y., Sun, P. T., Wu, X., Song, J. H., et al. (2017). Distinguishing Intracerebral Hemorrhage from Acute Cerebral Infarction through Metabolomics. Revista de Investigación Clínica - Clinical and Translational Investigation, 69, 319–328.
Zhang, Q.-S., & Zhu, S.-C. (2018). Visual interpretability for deep learning: A survey. Frontiers of Information Technology & Electronic Engineering, 19, 27–39.
Zhao, X. Y., Qin, W. J., & Qian, X. H. (2018). Application of deep learning in biological mass spectrometry and proteomics. Progress in Biochemistry and Biophysics, 45, 1214–1223.
Zheng, G., Zhang, F., Zheng, Z., Xiang, Y., Yuan, N. J., Xie, X., et al. (2018). DRN: A deep reinforcement learning framework for news recommendation. Proceedings of the 2018 World Wide Web Conference on World Wide Web, 1, 167–176.
Zou, J., Huss, M., Abid, A., Mohammadi, P., Torkamani, A., & Telenti, A. (2019). A primer on deep learning in genomics. Nature Genetics, 51, 12–18.
Zurada, J. M. (1992). Introduction to artificial neural systems. Minnesota: West Publishing Company.
Author information
Authors and Affiliations
Contributions
All authors conceived of the idea. KMM wrote the manuscript. DIB and SNR edited the manuscript.
Corresponding authors
Ethics declarations
Conflicts of interest
The authors have no disclosures of potential conflicts of interest related to the presented work.
Research involving human or animal rights
No research involving human or animal participants was performed in the construction of this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mendez, K.M., Broadhurst, D.I. & Reinke, S.N. The application of artificial neural networks in metabolomics: a historical perspective. Metabolomics 15, 142 (2019). https://doi.org/10.1007/s11306-019-1608-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-019-1608-0